Proteome
advertisement

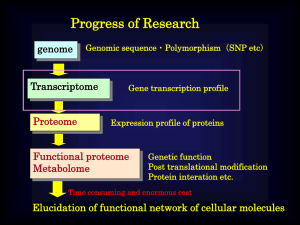
Genomic & Postgenomic Technologies Contents Introduction Gene diagnostics Transcriptome and its future direction Proteomics technologies Technologies for assessing protein-interaction Technologies for protein labeling Protein array and peptide array Mass spectroscopy & proteomics Monitoring of protein kinases In vivo imaging DDS & gene delivery http://www.chem.kyushu-u.ac.jp/~katayama/ Why do we need ‘Bio-Technologies’? 1. It makes great innovation & progress in our lives. Keeping good QOL and solving the issue of aging: Genomic drug discovery; Genomic diagnostics; New therapies; Regenerative Medicines Solving the issue of food supply: Improvement of food self-sufficiency Solving environmental issues and energy supply: Bio-process, bio-mass, bio-energy technologies 2. Growing Market in Bio-technologies Biotechnologies can make a new industrial field in worldwide. Market size:$ 2.3 trillion in 2010. Ex) Pharmaceutical industry Process of drug discovery Step of period The number of Candidates in the past 5 years in Japan development Basic 563, 589 compounds research 2~ 3 years Preparation & selection of new compounds Preclinical research of pharmacologic action, metabolic pathway, 3 ~ 5 years Examination 1/2790 202 compounds and safety of the selected compounds Devil’s river Clinical research Phase I : Confirmation of the safety for healthy persons 3 ~ 7 years Phase83IIcompounds : Research of safe administration 1/6790for patients Phase III: Research of effectiveness & safety or comparison Death with other drugs valley approval 1 ~ 2 years postcommercializing research During a sales period 35 compounds 1/16103 Research of safety, effectiveness and quality of! 1/21677 commercialized drug and promoting the appropriate use 26 compounds (1) R&D : 9~17 years (2) Cost : about $ 6,000 million~ $ 1 billion (3) Success rate : 1/21,677 the number of approved medicines in past 5 years in japan: 26 Characteristics if Biotechnology 1: Basic researches directly linked to applied researches All technologies are in developing Industry-academia cooperation is crucial. 2: Long incubation time Leading time is so long for practical application Product life time in drugs: 16 years to 9 years in each drug in this decade Leading time: 9 years to 13 years Development time is longer than the product lifetime! Industry-Academia relationship to acceleration of development Ideas & Speed! Whole industrial system will changed by using genomic information Current situation of genomic drug discovery Decoding of human genome Possible to find & use of disease-associated genes Although sequences of all human genes have been elucidated…. ・Most of drug target genes are still unknown. What is important to dicover drug target genes? Elucidation of the functions in diseases will be the key! Key point Important thing is to establish technologies that can evaluate the pathological roles of genes screened by medicinal research. From mechanism-oriented to disease-oriented Key issue that we have to establish will be… How we can validate the function of gene quickly? Establishment of new validation system of genetic function in vtro & in vivo. Current genomic researches have tried pulling out of all nails on the chest. However, the number of the nails may be infiinite… Even if we get the treasure chest (target gene), we can’t open it (because we can’t access to its function in disease.) We have got the map (genomic sequence) to find treasure so that we can get treasure chest. Key to ope the chast (Post genomic technology) Getting treasure (new drugs) !! Genomic research What is the difference between human & ape? Progress of Research genome Genomic sequence・Polymorphism(SNP etc) Transcriptome Proteome Gene transcription profile Expression profile of proteins Functional proteome Metabolome Genetic function Post translational modification Protein interation etc. Time consuming and enormous cost Elucidation of functional network of cellular molecules Technologies in each categories Genome ・DNA chip ・Invader assay ・Sniper assay ・PROBE assay ・Luminex ・PCR-SSCP ・PCR-RFLP etc Structure analysis(sequence) Polymorphism analysis Transcriptome ・cDNA chip etc ・Differential expression analysis Proteome Identification Protein function Protein interaction Ligand interaction Post translational modification ・protein chip, peptide chip ・Y2H ・SELEX ・Phage display ・STABLE assay etc Progress of research genome Genomic sequence・Polymorphism(SNP etc) Transcriptome Proteome Gene transcription profile Expression profile of proteins Functional proteome Metabolome Genetic function Post translational modification Protein interation etc. Time consuming and enormous cost Elucidation of functional network of cellular molecules Nucleic acid:DNA, RNA(mRNA, tRNA, rRNA) Missions of gene 1:Menteinance of genetic information:repairing 2:Transmission of genetic information:replication 3:Use of genetic information:transcription & translation Gene: Region of genomic DNA coding protein Genome : Whole set of genes in particular species Total gene is only 3% of whole genomic DNA Analysis of gene polymorphism Polymorphism marker:Difference of DNA sequence on the genome High polymorphism, but the distribution is less and heterogenious Mini-satellite:Repeat of several to tens of base sequence Micro-satellite:Repeat of 1 to 4 base sequence Base insertion and deletion: Insertion /Deletion of 1-tens of base sequence Low polymorphism, but are a lot of distributed on genomic DNA uniformly Single base polymorphism(SNP):1 /1000 bases, 3-10 millions SNA on human genome Why gene typing is needed? If gene type is elucidated, effectiveness or adverse effect of particular drug can be validated. In USA in 1994, 2 million people got extension of hospital stay and 100,000 people died due to the drug side effect. Medical expenses: $ 84 billion Cohort study of SNP mapping Social issues: Informed consent, Handling of data to protect personal information Intellectual property Technical issues Cost: Current technology takes $ 40 billion for the analysis of 1000 SNPs. Profiling of large number of SNPs is required for disease diagnostics. SNP analysis Identification & Mapping of SNPs Ability to find many SNPs from small number of genomic samples. SNPs Map SNPs Typing Ability to typing of particular (small amount of) SNPs by using a large number of genomoc samples If the SNPs typing is performed genome-wide, around 100 million of SNPs have to be typed. Speed & Cost Effectiveness! Allele specific hybridization Mini-sequencing Ligation assay Ligation with enzyme Ligation with enzyme G G A A ( A -A llele) ( G -A llele) A m plified D N A frag m ent ( A -A llele) A m plified D N A fragm ent ( G -A llele) F luorescein-labeled O D N C R O X -labeled O D N U F lu orescein-labeled O D N C T A M R A -labeled O D N U ROX-d d C TAMRA- d d U P C R prim ers G FRET FRET C G A U A FRET R O X positive C G T A M R A positive R O X p ositive FRET Endogenious SNPs typing using FRET a) TDI assay, b) DOL assay U G T A M R A p ositive b) a) c) G G A m p lified G -A llele R N A A m p lified G -A llele G h yb rid iz a tio n h yb rid iz a tion A m p lified G -A llele p r i me r p r ob e DNA p r i me r Ta g C TG CG G f l u or e s c e i n - d d CTP T p rim er array p rim er array b i ot i n - d d TTP G C sing le b ase ex tention C f l uor e s c e i n- d d CTP T b i ot i n -d d TTP C f l u or e s c e i n - d CTP d NTP sing le b ase ex tention C c Ta g CG olig o T ag -array TG CG p rim er ex tention SNPs typing using primer extention on a chip a)Oligo-Tag array, b) Primer array with single-base extention c)Primer array with multi-base extenton FRET b) a) flu o re p ho re T C FRET prim er C G q u encher com p lem entry seq u nece to the tem plate inclu d ing S N P M olecu lar B eacon T a q -polym e ra se P C R reaction G P C R reaction C C f l uor es c ence C G C G G C G F R E T probe is decom posed w ith the endonuclease activity F lu op rescence is increased w ith the P C R reaction SNPs typing using kinetic –PCR strategy a) Taq-Man PCR, b) Allele-specific molecular beacon Invader Assay Flap Flap Inveder probes EndoflapNNuclease Reporter probes C Cleavage G C N G Fluorophore 1 Quencher Cleavage N T Cleavage T N A A Fluorophore 2 Quencher Invader assay Flap Flap Invader probe cleavage Reporter probe C N G Fluorophore cleavage Quencher cleavage Invader probe T N A Fluorophore Reporter probe Quencher cleavage Advantage: PCR is unnecessary Drawback: Quite large amount of sample is required Background reaction exists Sniper assay Circular PCR + DNA sample containing SNP site Molecular beacon Cyclization Padlock probe Non-cyclization Luminex Assay PCR amplified DNA Zip code Capture probe ligation Reporter probe Cell sorter C15~C18 Fluorescent bead c-Zip code 25~20base Linker sequence 25~20base Pyro-sequencing SNP typing using Mass Spectrometry RFLP :restriction fragment length polymorphysm AATGATG TTACTAC AATGCTG TTACGAC AATG TTA SSOP AATGATG TTACTAC CTG CGAC :sequence specific oligonucleotide probe AATGCT TTACGA AATGCT TTACGA Biotin PCR amplified DNA Magnetic bead modified with streptavidin SNP typing using MS PINPOIN assay SNP typing using MS PROBE Assay SNP typing using MS VSET assay Survivor assay G + A A m p lif ied D N A re g io n in c lu d in g th e S N P s ite p rim e r h y b rid iz atio n G A ddA TP, ddGT P ddC TP, ddTTP e x ten tio n r ea ctio n G C A T A G C T h etero z y g o te m /e E S I- M S Schematic outline of Survivor assay The figure shows the case of heteroxygote.