Automated Microbial Genome Annotation
advertisement

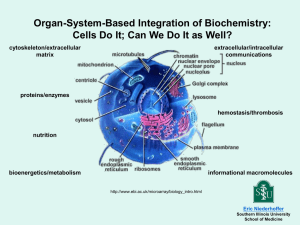
MICROBIAL GENOME ANNOTATION Loren Hauser Miriam Land Yun-Juan Chang Frank Larimer Doug Hyatt Cynthia Jeffries 1 NEB Educational Support http://www.neb.com/nebecomm/course_support.asp? 2 Why study Computational Biology and Bioinformatics? DNA sequencing output is growing faster than Moore’s law! 1 Illumina sequencing machine = 0.5 Tbp/week There are hundreds of these and thousands of other sequencing machines around the world. New sequencing technology will conceivably allow sequencing a human genome for less than $1K in less than 1 day! 3 Why study Medical Bioinformatics? In the near future, most cancer diagnostics will involved DNA or RNA sequencing! In the near future, every baby born in the developed world will have their genome sequenced. Protecting privacy and your doctors ability to use that information are the only real impediments! Hospitals are using DNA sequencing to track antibiotic resistant bacterial infections. 4 DOE Undergraduate Research in Microbial Genome Analysis and Functional Genomics http://www.jgi.doe.gov/education 5 Why Study Microbial Genomes? Large biological mass (50% of total) photosynthetic (Prochlorococcus) fix N2 gas to NH3 (Rhodopseudomonas) NH3 to NO2 (Nitrosomonas) bioremediation (Shewanella, Burkholderia) pathogens, BW (Yersinia pestis - plague) food production (Lactobacillus) CH4 production (Methanosarcina) H2 production (Rhodopseudomonas) 6 Example of Current Microbial Genome Projects UC Davis – FDA funded 100K bacterial genomes project associated with food. 5 years = 20K per year / 200 days/year = 100 genomes/day! 7 Web Resources and Contact Information http://genome.ornl.gov/microbial/ http://www.jgi.doe.gov/ http://genome.jgi-psf.org/ http://www.jcvi.org/ http://www.ncbi.nlm.nih.gov/ http://www.sanger.ac.uk/ http://www.ebi.ac.uk/ ftp://ftp.lsd.ornl.gov/pub/JGI artemis ready files for each scaffold = (feature table plus fasta sequence file) Contact: landml@ornl.gov; hauserlj@ornl.gov 8 9 Evolution of Sequencing Throughput Sequencing Technology Maxam and Gilbert Manual Sanger Automated Sanger (96 lanes/gel) Automated Sanger (384 capilaries) 454 sequencing (new titanium) Solexa (Illumina) Solexa (Illumina) PacBio realtime sequencing Samples/run bp/sample runs/week 1 100 5 5 400 5 100 500 5 400 600 10 1,000,000 400 5 300,000,000 75 1 1,000,000,000 200 1 100,000,000 1000 10 bp/week year 500 1977 10000 1985 250000 1995 2400000 2002 2E+09 2009 2.25E+10 2009 2.00E+12 ?2010 1E+12 ?2010 Sequenced Microbial Genomes ARCHAEAL GENOMES 159 FINISHED; 218 IN PROGRESS BACTERIAL GENOMES 3363 FINISHED; 11831 IN PROGRESS ENVIRONMENTAL COMMUNITIES > 50,000 samples (see MGRast) as of Sept 6, 2012 http://www.expasy.ch/alinks.html http://www.genomesonline.org http://metagenomics.anl.gov/ 11 Published Genomes Nitrosomonas europaea - J.Bac. 185(9):2759-2773 (2003) Prochlorococcus MED4 & MIT9313 - Nature 424:1042-1047 (2003) Synechococcus WH8102 - Nature 424:1037-1042 (2003) Rhodopseudomonas palustris - Nat. Biotech. 22(1):55-61 (2004) Yersinia pseudotuberculosis - PNAS 101(22):13826-31 (2004) Nitrobacter winogradskyi – Appl. Envir. Micro. 72(3):2050-63 (2006) Nitrosococcus oceani - Appl. Envir. Micro. 72(9):6299-315 (2006) Burkholderia xenovorans – PNAS 103(42):15280-7 (2006) Thiomicrospira crunogena – PLoS Biology 4(12):e383 (2006) Nitrosomonas eutropha C91 – Env. Micro. 9(12):2993-3007 (2007) Sulfuromonas denitrificans – Appl. Envir. Micro. 74(4):1145-56 (2008) Nitrosospira multiformis -- Appl. Envir. Micro. 74(11):3559-72 (2008) Nitrobacter hamburgensis -- Appl. Envir. Micro. 74(9):2852-63 (2008) Saccharophagus degradans – PLoS Genetics 4(5):e1000087 (2008) R. palustris – 5 strain comparison – PNAS 105(47):18543-8 (2008) L. rubarum and L. ferrodiazotrophum – Appl. Envir. Micro. (in press) 12 Basic Annotation Impacts Design of oligonucleotide arrays Design & prioritize protein expression constructs Design & prioritize gene knockouts Assessment of overall metabolic capacity Database for proteomics Allows visualization of whole genome 13 Additional Analysis Impacts Revised functional assignments based on domain fusions, functional clustering, phylogenetic profile Regulatory motif discovery Operon and regulon discovery Regulatory and protein association network discovery 14 Microbial Annotation Genome Pipeline Scaffolds or contigs Simple repeats Prodigal Complex Repeats Model correction tRNAs Final Gene List InterPro PRIAM Blast COGs TMHMM SignalP rRNA, Misc_RNAs GC Content, GC skew Function call Web Pages Feature table 15 Prodigal (Prokaryotic Dynamic Programming Genefinding Algorithm) Unsupervised: Automatically learns the statistical properties of the genome. Indifferent to GC Content: Prodigal performs well irrespective of the GC content of the organism. Draft: Prodigal can train on multiple sequences then analyze individual draft sequences. Open Source: Prodigal is freely available under the GPL. Reference: Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010 Mar 8;11(1):119. (Highly Accessed) 16 G+C Frame Plot Training Takes all ORFs above a specified length in the genome. Examines the G+C bias in each frame position of these ORFs. Does a dynamic programming algorithm using G+C frame bias as its coding scoring function to predict genes. Takes those predicted genes and gathers dicodon usage statistics. 17 Gene Prediction Dicodon usage coding score Length factor added to coding score (GCcontent-dependent) Coding/noncoding thresholds sharpened (starts downstream of starts with higher coding get penalized by the difference). Dynamic programming to put genes together. Bonuses for operon distances, larger bonus for -1/-4 overlaps. Same strand overlap allowed (up to 60 bases). Opposite strand -->3'r 5'f<- allowed (up to 250 bases) 18 Start Site Scoring Shine Dalgarno Motif Examines initially predicted genes and gathers statistics on the starts (RBS motifs, ATG vs GTG vs TTG frequency) Moves starts based on these discoveries. Gathers statistics on the new set of starts and repeats this process until convergence (5-10 iterations). RBS motifs based on AGGAGG sequence, 3-6 base motifs, with one mismatch allowed in 5 base or longer motifs (e.g. GGTGG, or AGCAG). Does a final dynamic programming with the start scoring function. 19 Start Site Scoring Other Motifs If Shine-Dalgarno scoring is strong, use it – this accounts for ~85% of genomes. If Shine-Dalgarno scoring is weak, look for other motifs If a strong scoring motif is found, use it (example GGTG in A. pernix) If no strong scoring motif is found, use highest score of all found motifs (example – Crenarchaea, Tc and Tl start sites are the same, but internal operon genes use weak Shine-Dalgarno motifs) 20 Annotated Gene Prediction 21 Prodigal Scoring 22 Gene Prediction Problems – Pseudogenes 23 Pseudogenes – Internal deletion 24 Pseudogenes – Premature stop codon 25 Pseudogenes – N-terminal deletion 26 Pseudogenes – Transposon insertion 27 Pseudogenes – Multiple frameshifts 28 Pseudogenes – Premature Stop and Frameshift 29 Pseudogenes – Dead Start Codon 30 31 GENE PAGE 32 33 34 35 ORGANISM’S (PSYC) COGS LIST Contig Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Contig1 Gene 1 3 4 6 8 10 11 14 15 16 17 19 20 23 24 25 26 27 28 29 29 30 31 33 34 35 36 37 38 39 40 41 42 43 45 46 47 51 52 53 57 58 59 59 60 61 62 64 65 66 67 68 69 70 71 74 76 77 78 78 Num Prot ------------------------------------------------------------- Group I E S K R L COG COG1211 COG0137 COG1376 COG0583 COG0628 COG0593 J G COG0172 COG0021 L L G R P P COG0551 COG0507 COG0057 COG1451 COG0168 COG0569 M COG2885 M R R R COG1538 COG1538 COG2274 COG1566 C R Q P S R P R COG2010 COG3019 COG2132 COG3667 COG3544 COG0491 COG2217 COG1826 S G O J J M S O O C O O F R I R M S P P E L O COG1937 COG2814 COG0435 COG0261 COG0211 COG2834 COG4399 COG1138 COG0526 COG0526 COG3088 COG4235 COG0563 COG1949 COG1502 COG0790 COG1519 COG1385 COG1840 COG1178 COG3842 COG0188 COG0625 R T COG0515 COG0515 Gene Name COG Des c ription Sc ore E-Value Category 4-diphos phoc Is pD y tidy l-2-methy l-D-erithritol 86 9.00E-19 s y nthas Lipid e metabolis m Argininos uc ArgG c inate s y nthas e 565 1.00E-162 Amino ac id trans Unc harac teriz ErfK ed protein c ons erv 61 ed 4.00E-11 in bac teria Func tion unk now Trans c riptional Ly s R regulator 123 2.00E-29 Trans c ription Predic ted permeas PerM e 145 6.00E-36 General func tion ATPas e inv DnaA olved in DNA replic 104 ation 8.00E-24 initiation DNA replic ation No COG Sery l-tRNASerS s y nthetas e 557 1.00E-160 Trans lation ribos Trans k etolas Tk e tA 992 0 Carbohy drate tra No COG Zn-finger domain TopA as s oc iated with 38 topois 9.00E-04 omeras DNA e ty replic pe I ation ATP-dependent Rec D ex oDNAs e (ex 61 onuc 3.00E-10 leas e V) DNA alphareplic s ubunit ation - h Gly c eraldehy GapA de-3-phos phate 344 dehy drogenas 7.00E-96e/ery Carbohy thros drate e-4-pho tra Predic ted metal-dependent COG1451 hy 104 drolas 3.00E-24 e General func tion Trk -ty pe K+ Trk trans G port s y s tems 181 membrane 1.00E-46 c omponents Inorganic ion tra K+ trans port Trk sA y s tems NAD-binding 125 4.00E-30 c omponent Inorganic ion tra No COG Outer membrane OmpA protein and 113 related 1.00E-26 peptidogly Cell c an-as envelope s oc iate bio No COG Outer membrane TolC protein 114 2.00E-26 Cell envelope bio Outer membrane TolC protein 114 2.00E-26 Cell envelope bio ABC-ty pe bac SunT terioc in/lantibiotic 410ex 1.00E-115 porters c ontain Defens an e mec N-termi han Multidrug res EmrA is tanc e efflux pump 82 7.00E-17 Defens e mec han No COG No COG Cy toc hrome Cc ccmonoA and diheme 38 v 3.00E-04 ariants Energy produc tio Predic ted metal-binding COG3019 protein 136 1.00E-33 General func tion Putative multic SufIopper ox idas es 271 9.00E-74 Sec ondary meta Unc harac teriz Pc ed oB protein involv 148 ed in 6.00E-37 c opper res Inorganic is tanc e ion tra Unc harac teriz COG3544 ed protein c ons erv 43 ed 8.00E-06 in bac teria Func tion unk now Zn-dependent GloB hy drolas es inc luding 100 2.00E-22 gly ox y lasGeneral es func tion Cation trans Zport ntA ATPas e 754 0 Inorganic ion tra Sec -independent TatA protein s ec retion 59 4.00E-11 pathway c Intrac omponents ellular traffi No COG Unc harac teriz COG1937 ed protein c ons erv 49 ed 5.00E-08 in bac teria Func tion unk now Arabinos e efflux AraJ permeas e 57 2.00E-09 Carbohy drate tra Predic ted glutathione ECM4 S-trans507 feras 1.00E-145 e Pos ttrans lationa Ribos omal RplU protein L21 126 3.00E-31 Trans lation ribos Ribos omal RpmA protein L27 119 4.00E-29 Trans lation ribos Outer membrane LolA lipoprotein-s 122 orting 3.00E-29 protein Cell envelope bio Unc harac teriz COG4399 ed protein c ons erv 41 ed 2.00E-04 in bac teria Func tion unk now Cy toc hrome Cc cmF biogenes is fac589 tor 1.00E-169 Pos ttrans lationa Thiol-dis ulfide Trx is A omeras e and thioredox 49 2.00E-07 ins Pos ttrans lationa Thiol-dis ulfide Trx is A omeras e and thioredox 49 2.00E-07 ins Pos ttrans lationa Unc harac teriz Cc ed mH protein involv 157 ed in 4.00E-40 bios y nthes Pos is ttrans of c -ty lationa pe c y t Cy toc hrome COG4235 c biogenes is fac103 tor 3.00E-23 Pos ttrans lationa Adeny late k Adk inas e and related168 k inas 3.00E-43 es Nuc leotide trans Oligoribonuc Orn leas e (3'->5' ex oribonuc 293 7.00E-81 leas e) RNA proc es s ing Phos phatidy Cls ls erine/phos phatidy 230 lgly 1.00E-61 c erophos Lipid phate/c metabolis ardiolipin m FOG: TPR COG0790 repeat SEL1 s ubfamily 56 4.00E-09 General func tion 3-deox y -D-manno-oc KdtA tulos onic 330 -ac id2.00E-91 trans feras Cell e envelope bio Unc harac teriz COG1385 ed protein c ons120 erved 1.00E-28 in bac teria Func tion unk now ABC-ty pe Fe3+ AfuA trans port s y s 164 tem periplas 1.00E-41 mic Inorganic c omponent ion tra ABC-ty pe Fe3+ ThiP trans port s y s 287 tem permeas 1.00E-78 e Inorganic c omponent ion tra ABC-ty pe s PotA permidine/putres c 318 ine trans 5.00E-88 port s y Amino s tems ac AT id Pas trans e Ty pe IIA topois Gy rA omeras e (DNA 941 gy ras e/topo 0 II DNA topois replic omeras ation e I GlutathioneGs S-trans t feras e 94 7.00E-21 Pos ttrans lationa No COG Serine/threonine SPS1protein k inas e 83 2.00E-17 General func tion Serine/threonine SPS1protein k inas e 83 2.00E-17 General func tion 36 Taxonomic Distribution of Top KEGG BLAST Hits 37 Frequency distance distributions Salgado et al. PNAS (2000) 97:6652 Fig. 2 38 Frequency distance distributions Salgado et al. PNAS (2000) 97:6652 Fig. 3b 39 Branched Chain Amino Acid Transporter family Organism Nostoc punctiforme Trichodesmium erythraeum Helicobacter pylori J99 Helicobacter pylori 26695 Campylobacter jejuni subsp. jejuni NCTC 11168 Geobacter metallidurans Desulfovibrio desulfuricans Escherichia coli K12 Escherichia coli O157:H7 EDL933 Buchnera sp. APS Pseudomonas aeruginosa, PAO1 Pseudomonas fluorescens Pseudomonas syringae Psychrobacter Vibrio cholerae O1 biovar eltor str. N16961 Yersinia pestis, CO92 Yersinia pseudotuberculosis Haemophilus influenzae Rd KW20 Pasteurella multocida subsp. multocida str. Pm70 Xylella fastidiosa (3 strains) Azotobacter vinlandii Psychrobacter Burkholderia fungorum Burkholderia mallei Burkhoderia pseudomallei Ralstonia metallidurans Ralstonia eutropha Nitrosomonas europaea Neisseria meningitidis MC58 Neisseria meningitidis Z2491 Caulobacter crescentus Mesorhizobium loti Agrobacerium tumefaciens Bradyrhizobium japonicum Brucella melitenis Brucella suis Sinorhizobium meliloti Rickettsia conorii Rickettsia prowazekii Rhodobacter sphaerodes Rhodospirillum rubrum Rhodopseudomonas palustris Cyano Cyano epsilon epsilon epsilon delta delta gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma gamma beta beta beta beta beta beta beta beta alpha alpha alpha alpha alpha alpha alpha alpha alpha alpha alpha alpha JGI JGI COG COG COG JGI JGI COG COG COG COG JGI JGI JGI COG COG JGI COG COG COG, JGI JGI JGI JGI JGI JGI JGI COG COG COG COG COG COG JGI JGI JGI ATPase ATPase Permease PBP COG0410 COG0411 COG0559 COG0683 3 3 6 4 2 1 3 6 0 0 0 0 0 0 0 0 1 1 2 2 1 1 2 1 2 2 4 4 1 1 2 2 1 1 2 2 0 0 0 0 3 3 7 4 3 3 5 3 7 4 8 5 0 0 1 0 0 0 1 0 2 2 4 2 2 2 4 2 0 0 0 0 0 0 0 0 0 0 0 0 3 4 8 2 0 0 0 0 22 20 34 29 6 6 11 8 7 7 13 10 9 8 16 12 18 19 36 28 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 7 7 16 10 7 7 15 9 27 26 50 59 7 7 12 13 6 6 12 11 5 5 12 8 0 0 1 0 0 0 1 0 6 6 12 5 6 7 13 9 20 20 40 38 40 Probable Ancient Gene (Liv Operon) 41 Branched Chain Amino Acid Transporter family – Rhodopseudomonas palustris Target ID Description Putative Ligand RPA0985 putative branched-chain amino acid transport system substrate-binding protein branched chain AAs RPA4029 RPA4648 possible branched-chain amino acid ABC transport system substrate-binding protein possible ABC transporter binding protein component branched chain AAs Thermal Shift Assay Binding Ligand 4-Hydroxybenzoate, Benzoate, Salicylate, Benzaldehyde 4-Hydroxybenzoate, pCoumarate Δ Tm °C for 1000 uM Ligand OR (100uM ) Ligand Tm(°C) No Ligand 29.0,13.5, 2.5, 2.0 56.5 17.0, 2.0 58.6 spermidine/putrescine p-Coumarate 2.0 55.5 RPA1250 amide-urea binding protein branched chain AAs Urea 5.0 63.0 RPA1789 putative branched-chain amino acid transport system substrate-binding protein branched chain AAs p-Coumarate 7.0 67.0 branched chain AAs Urea 6.0 59.5 branched chain AAs Ala, Gly,Ser, Met, Leu, Cys 11.5, 6.5, 4.5, 2.5, 2.0, 2.0 77.5 nitrate/taurine Malate 4.0 52.5 amino acids, prefers polar aas Met, Cys, His 10.0, 6.5, 3.5 63.0 13.0, (6.0, 2.0 ) 61.5 6.0, 3.0, 3.0, 2.0, 2.0 52.0 RPA3669 RPA3810 RPA2043 RPA2628 RPA0668 RPA1741 RPA2193 RPA3486 RPA2499 putative urea short-chain amide or branched-chain amino acid uptake ABC transporter periplasmic solute-binding protein precursor putative periplasmic binding protein of ABC transporter putative ABC transporter, periplasmic substratebinding protein polar amino acid ABC transport substrate-binding protein, aapJ-2 (aapJ-2) putative ABC transporter subunit, substrate-binding component possible branched-chain amino acid transport system substrate-binding protein putative ABC transporter, perplasmic binding protein, branched chain amino acids putative branched-chain amino acid transport system substrate-binding protein possible ABC transporter, periplasmic protein branched chain AAs branched chain AAs 4-Hydroxybenzoate, Salicylate, Benzaldehyde Met, Leu, Malate, Gly, Pro branched chain AAs Glutarate 5.0 64.5 branched chain AAs Glutarate 3.0 44.5 nitrate/taurine or aliphatic sulfonates Asn 7.0 53.5 42 Example of Lateral Transfer 43 Transporter Gene Loss in Yersina Pestis 36 Genes involved in transport from YPSE are nonfunctional in YPES 13 lost due to frameshifts 11 lost due to deletions 6 lost due to IS element insertions 4 (2 pair) lost due to recombination causing deletions and frameshifts 2 lost due to premature stop codons 44 45 Nostoc punctiforme Signal Transduction Histidine Kinases 46 Nostoc punctiforme Signal Transduction Histidine Kinases 47 Nostoc punctiforme Signal Transduction Histidine Kinases Gene # R1448 R1449 R1550 R1597 R1685 R1759 R1760 R1778 R1798 R1868 R2035 R2209 R2262 R2263 R2268 R2271 R2272 R2375 R2408 R2421 R2485 R2901 R2903 R2909 R3010 R3052 aa# 374 444 595 1042 1559 706 595 451 1098 713 1080 430 657 740 709 504 1801 1211 928 421 530 629 1116 103 210 475 COG COG0642 COG0642 COG0642 COG4191 COG0642 COG4251 COG0642 COG5002 COG0642 COG0642 COG0642 COG0642 COG0642 COG0642 COG0642 COG4191 COG3899 COG5278 COG4585 COG4585 COG0642 COG0642 COG4251 COG4251 COG0642 COG2205 N-term. (TM) N-term. RRR 1 1 unk. (2) Other domain PAS/PAC GAF(PHY) 1 Chase/1 Hpt unk. (2) unk.(1) unk. (3) 1 HAMP + TM 1 4 (3) 3(1) 2 (1) 1 1 1 5 1 1 1 1 2 1 1 Prt. Kin. 1 Chase 1 Cache (sp) unk. (1) unk. (4) unk. 1 1 unk. (1) 1 1 1 3 1 1 2 (1) 1 HisKA HATPase 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 1 1 1 1 1? 1 1 1 HisKA_3 1 HisKA_3 1 1 1 1 1 1 1 0 0.5 1 0 1 1 C-term. RRR Operon structure K1448/K1449 K1448/K1449 2 1 1 1 1 1 2 RRR1757/RRR1758/K1759/K1760 RRR1757/RRR1758/K1759/K1760 K1778/WHTH1779 K-R1798/K-F1799/LuxR-F1800 K-R2035/cNMP-F2036 2262-9 2262-9 2262-9 K2271/PK&K2272 K2271/PK&K2272 3 LuxR2407/K2408 LuxR2420/K2421 1 K2901/RRR2902/K2903 K2901/RRR2902/K2903 48 Nostoc punctiforme Signal Transduction Histidine Kinases 169 12 3 154 2 1 12 3 3 6 3 1 23 15 66 46 61 59 21 64 82 3 1 1 1 2 5 8 TM RRR predicted genes total pseudogenes genes with sensors but no kinase domain (do these work with the genes with no sensor domains - not in the same operon) functional Signal Transduction Histidine Kinases with 2 kinase domains (fused genes? or a 1 gene cascade?) with an Adenylate Cyclase domain with a Ser/Thr Protein Kinase domain, a COG3899 domain, 1 or more GAF domains, and possibly other domains with Hpt domain with CBS domains with Chase domains with Cache domains with an Amino acid transporter as a sensor? domain with a N-terminal RRR domain with only a N-terminal RRR as a sensor? domain with 1 or more RRR domains (86 RRR domains) with 1 or more C-terminal RRR domains (ie. Hybrid kinases) with 1 or more PAS/PAC domains (147 PAS/PAC domains total) with 1 or more GAF or Phytochrome domains (96 total - 38 phytochromes) with HAMP domains (34 total) with unknown N-terminal sensor domains with multiple N-terminal sensor domains with no sensor domain (do these work with the genes with no kinase domains - not in the same operon) with large C-terminal unknown domain with N-terminal RRR & WHTH (fused genes?) cNMP binding sensor domain with HisKA_2 type dimerization/autophosphorylation domains with HisKA_3 type dimerization/autophosphorylation domains putative operons with common (bidirectional) promoter Transmembrane alpha helical domain response regulator receiver domain (Phospho accepting Asp containing domain) 49 Nostoc punctiforme Regulatory Proteins 570 Regulatory Proteins Comments/Pseudogenes 201 Transcription/Elongation/Termination Factors 14 9 0 0 2 2 1 Sigma Factors Cyanobacterial Sigma Factors Sigma-54 (RpoN) Sigma 32 (RpoH) Sigma 28 (Flagella/Sporulation) Sigma-24 (RpoE/FecI) (ECF subfamily) Unknown Sigma factor (ECF subfamily) 17 1 8 1 1 5 1 Anti/Anti-Anti Sigma Factors Anti-Sigma regulatory factor (Ser/Thr protein kinase and phosphatase) Anti-Sigma-factor antagonist (STAS) domain protein Anti-Sigma-factor antagonist (STAS) and sugar transfersase Predicted transmembrane transcriptional regulator (anti-sigma factor) Putative Anti-Sigma regulatory factor (Ser/Thr protein kinase) Sigma 54 modulation protein/ribosomal protein S30EA 3 1 1 1 Termination/Antitermination Factors NusA antitermination factor NusB antitermination factor NusG antitermination factor 2 sets of pseudogenes: pNPAR018 truncated by transposase; pNPAR022, 3, 4 ar 1 set of pseudogenes: NpR2325/6 S1 RNA binding domain:KH domain / RNA binding 0 Elongation Factors 0 GreA/GreB family elongation factors 167 3 1 2 1 1 6 1 1 1 5 1 5 Transcription factors Ferric uptake regulator (FUR) family Negative regulator of class I heat shock protein phage shock protein A, PspA Phosphate uptake regulator, PhoU Plasmid maintenance system antidote protein Predicted transcriptional regulator SOS-response transcriptional repressor, LexA Putative transcriptional acitvator, Baf Transcriptional Regulator, AbrB family Transcriptional Regulator, AraC family Transcriptional Regulator, AraC family with Methyltransferase activity Two Component Transcriptional Regulator, AraC family 4 different COGs 50 Burkholderia xenovorans Regulatory Proteins 946 Regulatory Proteins Comments 704 Transcription/Elongation/Termination Factors 22 4 2 2 1 12 1 Sigma Factors Sigma 70 (RpoD) Sigma-54 (RpoN) Sigma 32 (RpoH) Sigma 28 (Flagella/Sporulation) Sigma-24 (RpoE/FecI) (ECF subfamily) Unknown Sigma factor (ECF subfamily) 13 1 1 1 2 4 1 1 1 1 Anti/Anti-Anti Sigma Factors Anti Sigma-E protein, RseA, Burkholderiaceae specific Anti-Sigma regulatory factor (Ser/Thr protein kinase and phosphatase) Anti-Sigma(ECF) factor, ChrR Anti-Sigma-factor antagonist (STAS) domain protein Predicted transmembrane transcriptional regulator (anti-sigma factor) Putative Anti-Sigma regulatory factor (Ser/Thr protein kinase) Putative Anti-Sigma-28 factor, FlgM Putative Sigma E regulatory protein, MucB/RseB Sigma-54 modulation protein 6 1 2 1 1 1 Termination/Antitermination Factors transcription termination factor Rho Response regulator receiver (CheY) and ANTAR domain protein NusA antitermination factor NusB antitermination factor NusG antitermination factor also called ribosomal protein S30AE Cold-shock DNA-binding domain(related to S1 RNA binding domain) ANTAR = RNA binding, anti-termination S1 RNA binding domain:KH domain / RNA binding 3 Elongation Factors 3 GreA/GreB family elongation factors 660 7 1 2 1 1 Transcription factors Cold-shock DNA-binding domain protein Possible Ferric uptake regulator (FUR) family Ferric uptake regulator (FUR) family Negative regulator of class I heat shock protein Negative transcriptional regulator 51 Regulatory Protein Identification Scheme Number Category Product Description COG1 COG2 InterPro COG0840 COG0840 Pfam Smart MCPsignal MA MCPsignal and Tar MCPsignal MCPsignal and Cache MCPsignal MCPsignal MA and TarH MA MA MA and GAF MA and GAF 5 5 5 5 5 5 5 5 5 Chemotaxis Signal Possible Transduction Bacterial chemotaxis sensory transducer Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer, TarH (aspartate) sensor Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer, Pas/Pac sensor Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer, Cache sensor Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer, GAF sensor Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer, Phytochrome sensor Chemotaxis Signal Bacterial Transduction chemotaxis sensory transducer, Phytochrome sensor 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 Chemotaxis Signal CheW Transduction protein COG0835 CheW Chemotaxis Signal CheW Transduction protein IPR002545 Chemotaxis Signal Two component Transduction CheW protein IPR002545 and IPR001789 Chemotaxis Signal Possible Transduction CheA Signal Transduction Histidine Kinases (STHK), weak homolog, COG0643 no good domain identification Chemotaxis Signal Possible Transduction CheA Signal Transduction Histidine Kinases (STHK) COG0643 HATPase_c Chemotaxis Signal CheA Signal Transduction Transduction Histidine Kinases (STHK) COG0643 IPR008207 and IPR003594 Chemotaxis Signal CheA Signal Transduction Transduction Histidine Kinases (STHK) IPR002545 and IPR003594 and IPR004105 Chemotaxis Signal CheB methylesterase Transduction COG2201 CheB_methylest Chemotaxis Signal CheB methylesterase Transduction IPR000673 and IPR001789 CheB_methylest Chemotaxis Signal Two component Transduction CheB methylesterase COG2201 IPR001789 CheB_methylest Chemotaxis Signal MCP methyltransferase, Transduction CheR-type COG1352 CheR Chemotaxis Signal MCP methyltransferase, Transduction CheR-type IPR000780 CheR Chemotaxis Signal MCP methyltransferase, Transduction CheR-type with PAS/PAC sensor COG1352 CheR Chemotaxis Signal MCP methyltransferase/methylesterase, Transduction CheR/CheB with PAS/PAC sensor COG1352 CheR and CheB_methylest Chemotaxis Signal CheC,Transduction inhibitor of MCP methylation COG1776 Chemotaxis Signal CheD,Transduction stimulates methylation of MCP proteins COG1871 TIGR IPR004089 COG0840 COG0840 COG0840 COG0840 COG0840 IPR001294 IPR001294 and IPR004089 sensory_box CheW CheW CheW HATPase_c HPT and CheW MeTrc MeTrc MeTrc MeTrc sensory_box sensory_box 52 Summary of automated transporter annotation --- Zymomonas 317 Transporter Proteins 69 82 116 2 2 14 29 1 2 3 4 5 8 9 Channels/Pores Electrochemical Potential-driven transporters Primary Active Transporters Group Translocators Transport Electron Carriers Accessory Factors Involved in Transport Incompletely Characterized Transport Systems 23 46 73 9 103 2 13 2 1 1 14 12 17 1.A 1.B 2.A 2.C 3.A 3.B 3.D 4.A 5.A 5.B 8.A 9.A 9.B alpha-type channels beta barrel porins Porters (uniporters, symporters, antiporters) Ion-gradient-driven energizers P-P-bond-hydrolysis-driven transporters Decarboxylation-driven transporters Oxidoreduction-driven transporters Phosphotransfer-driven group translocators Transmembrane 2-Electron Transfer Carriers Transmembrane 1-Electron Transfer Carriers Auxiliary transport proteins Recognized transporters of unknown biochemical mechanism Putative uncharacterized transport proteins 53 Zymomonas transporters complete listing GROUP Porters Porters 2.A.53 sulfate transporter or Xanthine/uracil/vitamin C transporter carbonic anhydrase, sulfate transporter SulP family 2 proteins or0489 2.A.53 or1027 2.A.53 GROUP Porters Porters Porters Porters Porters Porters Porters Porters 2.A.6 putative lipooligosaccharide nodulation factor exporter, NolGHI, RND superfamily hydrophobe/amphiphile efflux-1 HAE1, RND superfamily acriflavin resistance protein, RND superfamily efflux transporter, RND family, MFP subunit acriflavin resistance protein, RND superfamily acriflavin resistance protein, RND superfamily hopanoid biosynthesis associated RND transporter like protein HpnN export membrane protein SecD, RND superfamily 8 proteins or0146 or0252 or0704 or1290 or1378 or1379 or1439 or1719 GROUP Porters Porters Porters 2.A.64 twin-arginine translocation protein TatC twin-arginine translocation protein TatB twin-arginine translocation protein TatA 3 proteins or1107 2.A.64.1.1 or1108 2.A.64.1.1 or1109 2.A.64.1.1 GROUP Porters Porters Porters Porters Porters 2.A.66 multi antimicrobial extrusion protein MatE polysaccharide biosynthesis protein polysaccharide biosynthesis protein polysaccharide biosynthesis protein virulence factor MviN family 5 proteins or0190 or0202 or1191 or1303 or1478 GROUP Porters Porters 2.A.69 predicted transporter, putative auxin efflux carrier component predicted transporter, putative auxin efflux carrier component 2 proteins or0625 2.A.69.2./1 or0626 2.A.69.2./1 2.A.6.3/2 2.A.6.2 2.A.6.2 2.A.6.2/3.A.1.122/8.A.1 2.A.6.2 2.A.6.2 2.A.6.5/7 2.A.6.4.1 2.A.66.1 2.A.66.2 2.A.66.2 2.A.66.2 2.A.66.4.1 54 Transcriptome Analysis Pipeline: RNA sequences to GRN Collect RNAseq data Map reads to genomes Predict operons In silico Compare operon determinations (genome coordinates) Improve algorithm Determine orthologs with OrthoMCL Determine orthologous operons Calculate reads/bp Display frequency plot Cluster analysis of gene expression changes Determine operons from frequency plot Align orthologous promoters Determine TISs with 5’ RACE. Determine TFBS from alignments Cluster analysis from gene expression arrays GRN genetic regulatory network Predict TFBS In silico Dynamic range and sensitivity New gene, wrong start, riboswitch Small Regulatory RNA ??? Differential gene expression Operon with Internal Promoter 60 Long Term Vision Develop TPing SOPs, and an automated analysis pipeline. Initially produce TPs and preliminary GRNs for all important DOE microbial genomes (i.e. BESC), and eventually all DOE microbial genomes. Incorporate the TP analysis pipeline into ORNL’s automated microbial annotation pipeline, and eventually into IMG and GenBank files. Add additional experimental methods to improve the GRN determinations.