09_Bayesian_FIL2011Ma
advertisement

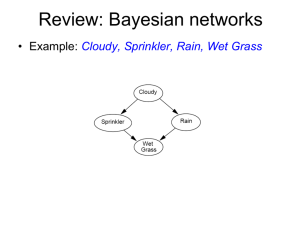
Bayesian inference J. Daunizeau Institute of Empirical Research in Economics, Zurich, Switzerland Brain and Spine Institute, Paris, France Overview of the talk 1 Probabilistic modelling and representation of uncertainty 1.1 Bayesian paradigm 1.2 Hierarchical models 1.3 Frequentist versus Bayesian inference 2 Numerical Bayesian inference methods 2.1 Sampling methods 2.2 Variational methods (ReML, EM, VB) 3 SPM applications 3.1 aMRI segmentation 3.2 Decoding of brain images 3.3 Model-based fMRI analysis (with spatial priors) 3.4 Dynamic causal modelling Overview of the talk 1 Probabilistic modelling and representation of uncertainty 1.1 Bayesian paradigm 1.2 Hierarchical models 1.3 Frequentist versus Bayesian inference 2 Numerical Bayesian inference methods 2.1 Sampling methods 2.2 Variational methods (ReML, EM, VB) 3 SPM applications 3.1 aMRI segmentation 3.2 Decoding of brain images 3.3 Model-based fMRI analysis (with spatial priors) 3.4 Dynamic causal modelling Bayesian paradigm probability theory: basics Degree of plausibility desiderata: - should be represented using real numbers - should conform with intuition - should be consistent • normalization: a=2 • marginalization: b=5 a=2 • conditioning : (Bayes rule) (D1) (D2) (D3) Bayesian paradigm deriving the likelihood function - Model of data with unknown parameters: y f - But data is noisy: e.g., GLM: f X y f - Assume noise/residuals is ‘small’: f 1 p exp 2 2 2 P 4 0.05 → Distribution of data, given fixed parameters: 2 1 p y exp 2 y f 2 Bayesian paradigm likelihood, priors and the model evidence Likelihood: Prior: Bayes rule: generative model m Bayesian paradigm forward and inverse problems forward problem p y , m likelihood posterior distribution p y , m inverse problem Bayesian paradigm model comparison Principle of parsimony : « plurality should not be assumed without necessity » y = f(x) Model evidence: y=f(x) x model evidence p(y|m) “Occam’s razor” : space of all data sets Hierarchical models principle ••• hierarchy causality Hierarchical models directed acyclic graphs (DAGs) Hierarchical models univariate linear hierarchical model prior densities posterior densities ••• Frequentist versus Bayesian inference a (quick) note on hypothesis testing • define the null, e.g.: H0 : 0 • invert model (obtain posterior pdf) p y p t H 0 P H0 y P t t * H 0 t t Y t* • estimate parameters (obtain test stat.) • define the null, e.g.: H0 : 0 • apply decision rule, i.e.: • apply decision rule, i.e.: if P t t * H 0 then reject H0 classical SPM if P H 0 y then accept H0 Bayesian PPM Frequentist versus Bayesian inference what about bilateral tests? • define the null and the alternative hypothesis in terms of priors, e.g.: p Y H 0 1 if 0 H 0 : p H 0 0 otherwise p Y H1 H1 : p H1 N 0, • apply decision rule, i.e.: if P H0 y P H1 y y 1 then reject H0 • Savage-Dickey ratios (nested models, i.i.d. priors): p y H 0 p y H1 p 0 y, H1 p 0 H1 Y space of all datasets Overview of the talk 1 Probabilistic modelling and representation of uncertainty 1.1 Bayesian paradigm 1.2 Hierarchical models 1.3 Frequentist versus Bayesian inference 2 Numerical Bayesian inference methods 2.1 Sampling methods 2.2 Variational methods (ReML, EM, VB) 3 SPM applications 3.1 aMRI segmentation 3.2 Decoding of brain images 3.3 Model-based fMRI analysis (with spatial priors) 3.4 Dynamic causal modelling Sampling methods MCMC example: Gibbs sampling Variational methods VB / EM / ReML → VB : maximize the free energy F(q) w.r.t. the “variational” posterior q(θ) under some (e.g., mean field, Laplace) approximation p 1 , 2 y , m p 1 or 2 y , m q 1 or 2 2 1 Overview of the talk 1 Probabilistic modelling and representation of uncertainty 1.1 Bayesian paradigm 1.2 Hierarchical models 1.3 Frequentist versus Bayesian inference 2 Numerical Bayesian inference methods 2.1 Sampling methods 2.2 Variational methods (ReML, EM, VB) 3 SPM applications 3.1 aMRI segmentation 3.2 Decoding of brain images 3.3 Model-based fMRI analysis (with spatial priors) 3.4 Dynamic causal modelling segmentation and normalisation realignment posterior probability maps (PPMs) smoothing dynamic causal modelling general linear model statistical inference normalisation p <0.05 template multivariate decoding Gaussian field theory aMRI segmentation mixture of Gaussians (MoG) model class variances 1 2 1 … k 2 yi … ith voxel label ith voxel value ci class frequencies k class means grey matter white matter CSF Decoding of brain images recognizing brain states from fMRI fixation cross + pace response >> log-evidence of X-Y sparse mappings: effect of lateralization log-evidence of X-Y bilateral mappings: effect of spatial deployment fMRI time series analysis spatial priors and model comparison PPM: regions best explained by short-term memory model short-term memory design matrix (X) prior variance of GLM coeff prior variance of data noise AR coeff (correlated noise) GLM coeff fMRI time series PPM: regions best explained by long-term memory model long-term memory design matrix (X) Dynamic Causal Modelling network structure identification m1 m2 m3 m4 attention attention attention attention PPC stim V1 V5 PPC stim V1 V5 PPC stim V1 V5 PPC stim V1 attention models marginal likelihood ln p y m 15 PPC 10 1.25 stim 5 estimated effective synaptic strengths for best model (m4) 0.10 0.26 V1 0.39 0.26 0.13 0.46 0 m1 m2 m3 m4 V5 V5 DCMs and DAGs a note on causality 1 1 2 2 21 3 1 1 3 2 time 2 32 13 3 t t 3 t 0 t t t x f ( x, u, ) 13u 3u u Dynamic Causal Modelling model comparison for group studies differences in log- model evidences ln p y m1 ln p y m2 m1 m2 subjects fixed effect assume all subjects correspond to the same model random effect assume different subjects might correspond to different models I thank you for your attention. A note on statistical significance lessons from the Neyman-Pearson lemma • Neyman-Pearson lemma: the likelihood ratio (or Bayes factor) test p y H1 p y H0 u is the most powerful test of size p u H 0 to test the null. • what is the threshold u, above which the Bayes factor test yields a error I rate of 5%? 1 - error II rate ROC analysis MVB (Bayes factor) u=1.09, power=56% CCA (F-statistics) F=2.20, power=20% error I rate