lect9
advertisement

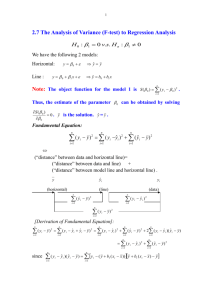
Lecture 9: ANOVA tables F-tests BMTRY 701 Biostatistical Methods II ANOVA Analysis of Variance Similar in derivation to ANOVA that is generalization of two-sample t-test Partitioning of variance into several parts • that due to the ‘model’: SSR • that due to ‘error’: SSE The sum of the two parts is the total sum of squares: SST 2.6 2.4 2.2 2.0 data$logLOS 2.8 3.0 Total Deviations: Yi Y 0 200 400 data$BEDS 600 800 2.6 2.4 2.2 2.0 data$logLOS 2.8 3.0 Regression Deviations: Yˆi Y 0 200 400 data$BEDS 600 800 2.6 2.4 2.2 2.0 data$logLOS 2.8 3.0 Error Deviations: Yi Yˆi 0 200 400 data$BEDS 600 800 Definitions Yi Y Yˆi Y Yi Yˆi SST (Yi Y ) 2 2 ˆ SSR (Yi Y ) 2 ˆ SSE (Yi Y ) SST SSR SSE Example: logLOS ~ BEDS > ybar <- mean(data$logLOS) > yhati <- reg$fitted.values > sst <- sum((data$logLOS- ybar)^2) > ssr <- sum((yhati - ybar )^2) > sse <- sum((data$logLOS - yhati)^2) > > sst [1] 3.547454 > ssr [1] 0.6401715 > sse [1] 2.907282 > sse+ssr [1] 3.547454 > Degrees of Freedom Degrees of freedom for SST: n - 1 • one df is lost because it is used to estimate mean Y Degrees of freedom for SSR: 1 • only one df because all estimates are based on same fitted regression line Degrees of freedom for SSE: n - 2 • two lost due to estimating regression line (slope and intercept) Mean Squares “Scaled” version of Sum of Squares Mean Square = SS/df MSR = SSR/1 MSE = SSE/(n-2) Notes: • mean squares are not additive! That is, MSR + MSE ≠SST/(n-1) • MSE is the same as we saw previously Standard ANOVA Table SS df MS Regression SSR 1 MSR Error SSE n-2 MSE Total SST n-1 ANOVA for logLOS ~ BEDS > anova(reg) Analysis of Variance Table Response: logLOS Df Sum Sq Mean Sq F value Pr(>F) BEDS 1 0.64017 0.64017 24.442 2.737e-06 *** Residuals 111 2.90728 0.02619 Inference? What is of interest and how do we interpret? We’d like to know if BEDS is related to logLOS. How do we do that using ANOVA table? We need to know the expected value of the MSR and MSE: E ( MSE) 2 E ( MSR) 2 12 ( X i X ) 2 Implications E ( MSE) 2 E ( MSR) 2 12 ( X i X ) 2 mean of sampling distribution of MSE is σ2 regardless of whether or not β1= 0 If β1= 0, E(MSE) = E(MSR) If β1≠ 0, E(MSE) < E(MSR) To test significance of β1, we can test if MSR and MSE are of the same magnitude. F-test Derived naturally from the arguments just made Hypotheses: • H0: β1= 0 • H1: β1≠ 0 Test statistic: F* = MSR/MSE Based on earlier argument we expect F* >1 if H1 is true. Implies one-sided test. F-test The distribution of F under the null has two sets of degrees of freedom (df) • numerator degrees of freedom • denominator degrees of freedom These correspond to the df as shown in the ANOVA table • numerator df = 1 • denominator df = n-2 Test is based on MSR F* ~ F (1, n 2) MSE Implementing the F-test The decision rule If F* > F(1-α; 1, n-2), then reject Ho If F* ≤ F(1-α; 1, n-2), then fail to reject Ho 0.8 F-distributions 0.4 0.2 0.0 f1 0.6 F(1,10) F(1,1000) F(5,10) F(5,1000) 0 1 2 3 x 4 5 6 ANOVA for logLOS ~ BEDS > anova(reg) Analysis of Variance Table Response: logLOS Df Sum Sq Mean Sq F value Pr(>F) BEDS 1 0.64017 0.64017 24.442 2.737e-06 *** Residuals 111 2.90728 0.02619 > qf(0.95, 1, 111) [1] 3.926607 > 1-pf(24.44,1,111) [1] 2.739016e-06 More interesting: MLR You can test that several coefficients are zero at the same time Otherwise, F-test gives the same result as a ttest That is: for testing the significance of ONE covariate in a linear regression model, an F-test and a t-test give the same result: • H0: β1= 0 • H1: β1≠ 0 general F testing approach Previous seems simple It is in this case, but can be generalized to be more useful Imagine more general test: • Ho: small model • Ha: large model Constraint: the small model must be ‘nested’ in the large model That is, the small model must be a ‘subset’ of the large model Example of ‘nested’ models Model 1: LOSi 0 1 INFRISK 2 MS 3 NURSE 4 NURSE2 ei Model 2: LOSi 0 1 INFRISK 3 NURSE 4 NURSE2 ei Model 3: LOSi 0 1 INFRISK 2 MS ei Models 2 and 3 are nested in Model 1 Model 2 is not nested in Model 3 Model 3 is not nested in Model 2 Testing: Models must be nested! To test Model 1 vs. Model 2 • we are testing that β2 = 0 • Ho: β2 = 0 vs. Ha: β2 ≠ 0 • If β2 = 0 , then we conclude that Model 2 is superior to Model 1 • That is, if we reject the null hypothesis Model 1: LOSi 0 1 INFRISK 2 MS 3 NURSE 4 NURSE2 ei Model 2: LOSi 0 1 INFRISK 3 NURSE 4 NURSE2 ei R reg1 <- lm(LOS ~ INFRISK + ms + NURSE + nurse2, data=data) reg2 <- lm(LOS ~ INFRISK + NURSE + nurse2, data=data) reg3 <- lm(LOS ~ INFRISK + ms, data=data) > anova(reg1) Analysis of Variance Table Response: LOS Df Sum Sq Mean Sq F value Pr(>F) INFRISK 1 116.446 116.446 45.4043 8.115e-10 *** ms 1 12.897 12.897 5.0288 0.02697 * NURSE 1 1.097 1.097 0.4277 0.51449 nurse2 1 1.789 1.789 0.6976 0.40543 Residuals 108 276.981 2.565 --- R > anova(reg2) Analysis of Variance Table Response: LOS Df Sum Sq Mean Sq F value Pr(>F) INFRISK 1 116.446 116.446 44.8865 9.507e-10 *** NURSE 1 8.212 8.212 3.1653 0.078 . nurse2 1 1.782 1.782 0.6870 0.409 Residuals 109 282.771 2.594 --- > anova(reg1, reg2) Analysis of Variance Table Model 1: Model 2: Res.Df 1 108 2 109 LOS ~ INFRISK + ms + NURSE + nurse2 LOS ~ INFRISK + NURSE + nurse2 RSS Df Sum of Sq F Pr(>F) 276.981 282.771 -1 -5.789 2.2574 0.1359 R > summary(reg1) Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 6.355e+00 5.266e-01 12.068 < 2e-16 *** INFRISK 6.289e-01 1.339e-01 4.696 7.86e-06 *** ms 7.829e-01 5.211e-01 1.502 0.136 NURSE 4.136e-03 4.093e-03 1.010 0.315 nurse2 -5.676e-06 6.796e-06 -0.835 0.405 --Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.601 on 108 degrees of freedom Multiple R-squared: 0.3231, Adjusted R-squared: 0.2981 F-statistic: 12.89 on 4 and 108 DF, p-value: 1.298e-08 > Testing more than two covariates To test Model 1 vs. Model 3 • we are testing that β3 = 0 AND β4 = 0 • Ho: β3 = β4 = 0 vs. Ha: β3 ≠ 0 or β4 ≠ 0 • If β3 = β4 = 0, then we conclude that Model 3 is superior to Model 1 • That is, if we reject the null hypothesis Model 1: LOSi 0 1 INFRISK 2 MS 3 NURSE 4 NURSE2 ei Model 3: LOSi 0 1 INFRISK 2 MS ei R > anova(reg3) Analysis of Variance Table Response: LOS Df Sum Sq Mean Sq F value Pr(>F) INFRISK 1 116.446 116.446 45.7683 6.724e-10 *** ms 1 12.897 12.897 5.0691 0.02634 * Residuals 110 279.867 2.544 --- > anova(reg1, reg3) Analysis of Variance Table Model 1: Model 2: Res.Df 1 108 2 110 LOS ~ INFRISK + ms + NURSE + nurse2 LOS ~ INFRISK + ms RSS Df Sum of Sq F Pr(>F) 276.981 279.867 -2 -2.886 0.5627 0.5713 R > summary(reg3) Call: lm(formula = LOS ~ INFRISK + ms, data = data) Residuals: Min 1Q Median -2.9037 -0.8739 -0.1142 3Q 0.5965 Max 8.5568 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 6.4547 0.5146 12.542 <2e-16 *** INFRISK 0.6998 0.1156 6.054 2e-08 *** ms 0.9717 0.4316 2.251 0.0263 * --Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ Residual standard error: 1.595 on 110 degrees of freedom Multiple R-squared: 0.3161, Adjusted R-squared: 0.3036 F-statistic: 25.42 on 2 and 110 DF, p-value: 8.42e-10 Testing multiple coefficients simultaneously Region: it is a ‘factor’ variable with 4 categories LOSi 0 1 I ( Ri 2) 2 I ( Ri 3) 3 I ( Ri 4) ei