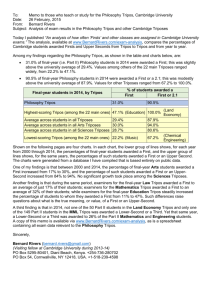
Computational Challenges of PK/PD NLME
Bob Leary
Pharsight Corporation
© Tripos, L.P. All Rights Reserved
Computational challenge #1 – make execution time
reasonable
•Many PK/PD NLME software packages - NONMEM (with many
choices for methods) is by far the most popular, but not necessarily
always the most appropriate
•All methods are to some degree computationally intensive –
execution time can be a limiting factor, even for a single run
• Many types of analyses require multiple runs (bootstrap,
covariate search, likelihood profiling, etc. – execution time
constraints can be severe).
© Tripos, L.P. All Rights Reserved
Slide 2
Execution time, cont’d
•There are trades-offs between accuracy/statistical quality and
speed: FO vs FOCE vs MCPEM/SAEM/NPAG
•Technology (parallel computing) can help a lot, but algorithmic
improvements are at least equally important (SAEM, MCPEM vs.
FOCE)
© Tripos, L.P. All Rights Reserved
Slide 3
Pipericillin model convergence with grid size
-400
-450
Log likelihood
-500
-550
-600
-650
-700
-750
-800
3
10
4
10
5
10
6
10
7
10
Number of grid points
© Tripos, L.P. All Rights Reserved
Slide 4
8
10
9
10
NPAG Outperforms NPEM
CPU HRS
MB
LOG -LIK
NPEM:
2037
10000
-433.1
NPAG:
0.5
6
-425.0
© Tripos, L.P. All Rights Reserved
Slide 5
Computational Challenges #2 - #4
•PK/PD NLME models and data are complex and computationally
demanding, probably much more so that most other NLME application
areas. Special purpose software is needed.
•Many of the methods are complex, not well documented,
approximate, not easily understood by the user base, and at least
somewhat fragile
•Software is relatively difficult to learn and use
© Tripos, L.P. All Rights Reserved
Slide 6
A chronology of events in development of NLME
1972 – Sheiner, Rosenberg, Melmon paper (FO)
1977 – NONMEM group established at UCSF
(L. Sheiner and S. Beal)
1979 – First NONMEM FO program appears
1986 – First nonparametric method NPML (A. Mallet)
1990 – First FOCE method (Lindstrom/Bates)
1990 – First Bayesian method (Gelfand/Smith – Bugs and PKBugs)
© Tripos, L.P. All Rights Reserved
Slide 7
Chronology, cont’d
1991 - NPEM nonparametric method (Schumitzky)
1992 – First PAGE meeting (63 participants, 500+ in 2010)
1993 - First Laplacian method - enables general LL models
(Wolfinger)
1999 – FDA Guidance for POP PK
2004 –2005 EM methods (SAEM, MCPEM, PEM) , Lyon inter-method
comparison exercises, MONOLIX
2007 – EMEA guidelines for POP PK
2009 – NONMEM SAEM/MCPEM/Bayesian, Pharsight PHOENIX
© Tripos, L.P. All Rights Reserved
Slide 8
Some PK/PD software
•NONMEM (L. Sheiner and S. Beal, UCSF 1979 – to date)
-primarily parametric modeling, although has primitive NP method
-classical approximate likelihood methods (FO, FOCE, FOCEI,
Laplacian)
-’new’ accurate likelihood EM methods (SAEM and MCPEM) (2009)
-Bayesian methods (2009)
•USC*PACK (R. Jelliffe, USC/LAPK et al., 1993-to date)
-nonparametric (NPEM, NPAG) (A. Schumitzky, R. Leary)
-individual dosing optimization – multiple model control (D. Bayard)
© Tripos, L.P. All Rights Reserved
Slide 9
PK/PD software, cont’d
•Monolix (INSERM, 2005 - to date) - SAEM (Stochastic Approximation
Expectation Maximization)
•Adapt/S-Adapt (USC/BMSR, D. D’Argenio, R. Bauer, 1989-to date)
MCPEM (Monte Carlo Parametric Expectation Maximization) + Bayesian
•PHOENIX (Pharsight, 2009 – to date) classical NM methods + AGQ +
SAEM + QMCPEM + NPAG + WinNonLin single subject and NCA modeling
•BUGS, WinBUGS – (1999 to date) – Bayesian
•S+ NLME, R NLME, SAS PROC-NLMIXED can be used, but not well
suited for PK/PD
© Tripos, L.P. All Rights Reserved
Slide 10
PK/PD Software User Base
WinNonLin (Single Subject, NCA): 6000 (3000 academic,
3000 commercial)
NONMEM (Population NLME): 1500
Commercial demand for experienced users exceeds supply
© Tripos, L.P. All Rights Reserved
Slide 11
FDA Guidance for Industry, 1999
Population PK analysis is concerned with identifying and quantifying
the random [random effects] and nonrandom [covariate effects]
variability in the PK behavior of the patient population
About 25% of recent submissions at time of writing included a
‘population’ analysis
Magnitude of random variability is particularly important because the
safety and efficacy of a drug is affected.
Mentions Standard Two Stage and NLME modeling as possible methods
© Tripos, L.P. All Rights Reserved
Slide 12
EMEA Guidelines 2007
NLME Pop PK analysis appears to be mandatory, or at least expected
No mention of STS
Extensive specification of model validation diagnostics and validation
techniques (CWRES, predictive checks, etc.)
Notes FDA Guidance is from 1999 and
“The FDA guidance should be read bearing in mind that it was written in
1999 and that population pharmacokinetics is an evolving science”
© Tripos, L.P. All Rights Reserved
Slide 13
Obligatory ODE section
© Tripos, L.P. All Rights Reserved
Slide 14
ODE Considerations
•Most PK models are dynamical systems that can be described by
ordinary differential equations (ODEs)
•ODEs often need to be solved numerically (many PK/PD software
packages use ODEPACK, a library of ODE solvers developed by A.
Hindmarsh at LLNL)
•If system is linear and homogeneous with constant coefficients, the
matrix exponential can be used
•Some special cases (1, 2, and 3-compartment models) are best
handled by built-in closed form solutions.
•Special handling capabilities are built in to the software for lag
times, bioavailability, etc.
© Tripos, L.P. All Rights Reserved
Slide 15
A Simple PK Model as ODE : 1-Compartment IV Bolus
dA / dt K A
C (t ) A(t ) / V
A(0) Dose
© Tripos, L.P. All Rights Reserved
Slide 16
IV Bolus closed form solution
Kt
e
C (t ) D
V
1
0.9
plasma concentration
0.8
0.7
0.6
0.5
0.4
0.3
0.2
0.1
t1/2=0.46
0
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
time t
© Tripos, L.P. All Rights Reserved
Slide 17
2
Multiple Doses: Use superposition if model ODE is linear
Kt
( Dose0 ) e
C (t )
, 0 t T1
V
( Dose0 ) e Kt ( Dose1 )e K (t T1 )
C (t )
, t T1
V
Covariate models with time varying covariates pose additional
complications – suppose K=tvK(1+(coef)(SCR-SCR0))
© Tripos, L.P. All Rights Reserved
Slide 18
1-Comp first order absorption extra-vascular dosing
d A1 / dt k12 A1
d A2 / dt k12 A1 k22 A2
C (t ) A2 (t ) / V
A1(0) Dose
k12 1st order absorption rate constant
k22 elimination rate constant
© Tripos, L.P. All Rights Reserved
Slide 19
1-Comp first order absorption extra-vascular dose
solution
1-compartment extravascular first order aborption
0.09
0.08
0.07
conc
0.06
0.05
0.04
0.03
0.02
0.01
0
0
0.1
0.2
0.3
0.4
0.5
time
0.6
0.7
0.8
0.9
1
D k12
C (t )
(e k22t e k12t )
V (k12 k22 )
© Tripos, L.P. All Rights Reserved
Slide 20
1 compartment 0-order (IV) dosing ODE
d A1 / dt 0
d A2 / dt k12 A1 k22 A2
A1(0) 1
A2 (0) 0
C (t ) A2 (t ) / V
k12 IV infusion rate
k22 elimination rate constant
© Tripos, L.P. All Rights Reserved
Slide 21
General N-compartment model : 0 and 1st order dosing
d A / dt K ij A
A(0) f (dosing at t=0)
General solution using matrix exponential
A(t ) e[ Kt ] A(0)
[ M ]2
e 1 M
...
2!
Accurate, fast, and reliable software libraries for matrix exponentials exist
and outperform numerical ODE solvers
M
© Tripos, L.P. All Rights Reserved
Slide 22
Nonlinear cases must be solved numerically with ODE
solvers (ODEPACK)
Michaelis-Menten elimination
V max
dA / dt
A
Km A
A(t )
C (t )
V
A(0) Dose
© Tripos, L.P. All Rights Reserved
Slide 23
ODE ‘solver’ order of preference/speed
1. Closed form (1, 2, 3 compartment, 0 and 1st order dosing)
2. Matrix Exponential (Linear, constant coefficient)
3. Non-stiff numerical ODE solver (Runge-Kutta, Adams)
4. Stiff ODE solver (Gear BDF)
Node execs = (Niter_out)(Nfix+Nran)(Nsub)(Niter_in)(Nran)(Ntime)
(100)(10)(1000)(20)(5)(10) = 1,000,000,000
© Tripos, L.P. All Rights Reserved
Slide 24
End of ODE section, Start of methods section
© Tripos, L.P. All Rights Reserved
Slide 25
Simple (single subject) regression Model
•PK Model
e Kt
C (t ) D
V
•Data
Concentration profile: (t j , Cobs (t j ) ), j 1,.., N obs
•Residual Error Model
Cobs (t ) C (t ) C (t )
~ N (0, 2 )
© Tripos, L.P. All Rights Reserved
Slide 26
Extended least squares objective function
ELS (V , K , 2 ) 2 ln(l (V , K , 2 )) const
Nobs
j 1
(Cobs (t j ) De
( De
Kt j
Kt j
/V ) 2
/V ) 2 2
ln( De
© Tripos, L.P. All Rights Reserved
Kt j
Slide 27
/V ) 2 2 )
Computational challenge : minimize ELS (V , K , )
2
•Nonlinear, nonconvex,
•But no likelihood approximations are necessary in single
subject case
•Unconstrained (can add bound constraints if desired)
•No exploitable structure
•Use general purpose unconstrained quasi-Newton method
UNCMIN from TOMS is 99+% reliable, but may encounter
problems with multiple minima
© Tripos, L.P. All Rights Reserved
Slide 28
Regression model to estimate V and K
0
10
log (V) = -log (C) - Kt
C(t)
slope = -K
V = 1/C(0)
-1
10
0
0.2
0.4
0.6
0.8
1
1.2
1.4
time t
© Tripos, L.P. All Rights Reserved
Slide 29
1.6
A simple population PK model: IV Bolus cont’d
V (tvV )eV or V etvlV V
K (tvK )eK
(V , K ) N (0, )
Data : (Cobs (tij ), tij , Di ), i 1, Nsub, j 1, N obsi
parameters to be fit:
fixed effects: tvV , tvK
residual error: 2
population covariance elements: VV , VV , KV
© Tripos, L.P. All Rights Reserved
Slide 30
Population Likelihood function
log L
Nsub
log( L )
i 1
i
Li (tvV , tvK , 2 , )
2
l
(
tvV
,
tvK
,
| V , K )h(V , K | ) dV d K )
i
J ( | tvV , tvK , 2 , )d
© Tripos, L.P. All Rights Reserved
Slide 31
Li cannot be evaluated analytically – how to proceed?
•Numerical quadrature - adaptive Gaussian quadrature, Monte Carlo
integration , quasi-Monte Carlo integration – very slow,
dimensionality problems
• Laplace approximation – FO, FOCE, Laplace (Y. Wang, 2006)
•Use a method that does not require integration (SAEM,PEM, MCPEM,
Bayesian methods, nonparametric methods)
© Tripos, L.P. All Rights Reserved
Slide 32
Laplacian Approximation (FO, FOCE, Laplacian)
J ( ) Ae
( )' H ( )
2
d /2
J
(
)
d
A
(2
)
/ det( H )
A J ( mode )
2 J ( mode )
H
2
© Tripos, L.P. All Rights Reserved
Slide 33
Joint log likelihood J(q,2,,) and Laplacian,
FOCE, and FO approximations
Joint likelihood and Laplace, FOCE, FO approximations
1.8
1.6
1.4
1.2
1
J(eta)
FO
0.8
FOCE
0.6
Laplace
0.4
0.2
0
-2
-1
0
1
2
3
eta
© Tripos, L.P. All Rights Reserved
Slide 34
4
5
Conditional methods (FOCE, Laplace) require nested
optimizations to find mode of J, FO does not
Each top level evaluation of
Nsub
log L log( Li )
i 1
requires Nsub mode-finding optimizations of
J ( | tvV , tvK , 2 , )d
Total number of innter optimizations = (Neval)(Nsub) - can
easily reach 100,000 or more, leading to a reliability problem
© Tripos, L.P. All Rights Reserved
Slide 35
Lyon 2004-2005 ‘bake-off’ of NLME methods
© Tripos, L.P. All Rights Reserved
Slide 36
© Tripos, L.P. All Rights Reserved
Slide 37
STATISTICAL EFFICIENCIES
© Tripos, L.P. All Rights Reserved
Slide 38
Approximate likelihoods can
destroy statistical efficiency
16
14
histogram (white)
of PEM estimators
12
histogram (blue) of
NONMEM FO
estimators
10
8
6
4
2
0
0.04
0.06
0.08
0.1
© Tripos, L.P. All Rights Reserved
0.12
0.14
Slide 39
0.16
0.18
SAEM, MCPEM, NPEM/NPAG
© Tripos, L.P. All Rights Reserved
Slide 40
The ideal case –Vi and Ki can be observed
Parametric estimators
Nonparametric histogram
1 N
ˆV Vi
N i 1
80
70
1 N
2
ˆ
ˆ
V
(Vi V )
( N 1) i 1
1/ 2
frequency
60
50
40
30
20
10
0
0.5
1
1.5
V
F {(Vi , Ki ), pi 1/ N}
© Tripos, L.P. All Rights Reserved
Slide 41
2
The real case: Vi and Ki are not directly
observable
We only have time profiles of drug
plasma concentrations
Measurement and dosing protocols
are not uniform over different
individuals
At best, we can get estimates
Vˆi , Kˆ i
by solving a regression model
© Tripos, L.P. All Rights Reserved
Slide 42
Standard Two-Stage Method Vi and Ki are estimated by
simple nonlinear regression methods
Parametric estimators
Nonparametric histogram
1 N
ˆV Vi
N i 1
80
70
1 N
2
ˆ
ˆ
V
(Vi V )
( N 1) i 1
1/2
frequency
60
50
40
30
20
10
0
0.5
1
1.5
V
F {(Vi , Ki ), pi 1/ N}
© Tripos, L.P. All Rights Reserved
Slide 43
2
MCPEM and SAEM are Monte Carlo versions of STS
1. inpute (Vik , K ik ), k 1, Nsamp for each subject i
by drawing random samples from the
(unnormalized) posterior:
(V , K ) ~ J i ( | tvV , tvK , 2 , )
( MCPEM : Nsamp ~ 500, SAEM: Nsamp ~ 1)
2. Compute ik 2 from inputed C(t) and data
3. Compute updated tvV , tvK , 2 , values
from STS formulas - no numerical optimization is necessary
© Tripos, L.P. All Rights Reserved
Slide 44
NPEM and NAG: Many PK/PD populations have subpopulations that would be missed by parametric techniques
A - True two-parameter population
distribution
B – Best normal approximation to
population distribution
© Tripos, L.P. All Rights Reserved
Slide 45
NPEM and NPAG
1. Assign an unknown probability (or probability density
value) pj to each grid point
2. Grid the relevant portion of the (V,K) with grid points
(Vj,Kj)
3. Estimate probabilities pj by maximizing the (exact)
nonparametric log likelihood
Nk
log LNP log( p j lij )
i 1
j
p j 0, p j 1
j
© Tripos, L.P. All Rights Reserved
Slide 46
NPEM vs NPAG
•NPEM uses a fixed, static grid and and EM algorithm to solve
optimization problem (no formal numerical optimization) for the
probabilities pj
•NPAG uses an adaptive grid (multiple iterations) and a convex special
purpose primal-dual algorithm to optimize the log likelihood
•A later extension of NPAG incorporated a d-optimal design criterion
based on the dual solution that enables candidate new grid points to
be tested very rapidly for potential for improving the likelihood
•Final optimal nonparametric distribution is discrete with at most
Nsub support points.
© Tripos, L.P. All Rights Reserved
Slide 47
NPAG results format looks like ideal case of direct observation
2.5
2
K
1.5
1
0.5
0
0
0.5
1
1.5
2
V
© Tripos, L.P. All Rights Reserved
Slide 48
2.5
PHX NPAG vs FOCE for bimodal distribution of
Ke values
35
60
30
50
25
40
20
30
15
20
10
10
5
0
0
-1
-0.8
-0.6
-0.4 -0.2
0
0.2
0.4
Simulated (true) Ke values
0.6
0.8
1
-1
-0.8
-0.6
-0.4 -0.2
0
0.2
0.4
Post-hoc estmate of eta Ke
90
80
70
60
50
40
30
20
10
0
-1
-0.8 -0.6 -0.4 -0.2
0
0.2
0.4
0.6
0.8
Nonparameteric mean eta Ke - optimized support points
© Tripos, L.P. All Rights Reserved
1
Slide 49
0.6
0.8
1