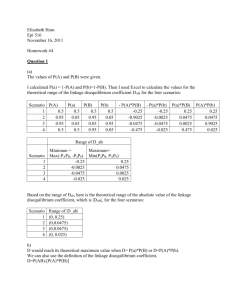
A a G - Tufts

Multiple Comparisons
Measures of LD
Jess Paulus, ScD
January 29, 2013
Today’s topics
1.
2.
Multiple comparisons
Measures of Linkage disequilibrium
• D’ and r 2
• r 2 and power
Multiple testing & significance thresholds
Concern about multiple testing
Standard thresholds (p<0.05) will lead to a large number of “significant” results
Vast majority of which are false positives
Various approaches to handling this statistically
Possible Errors in Statistical Inference
Reject
H
0
: SNP prevents
DM
Observed in the
Sample Fail to reject H
0
:
No assoc.
Unobserved Truth in the Population
H
0
: No
H a
: SNP prevents DM association
True positive
(1 – β)
False positive
Type I error (α)
False negative
Type II error ( β) :
True negative
(1α)
Probability of Errors
α
= Also known as: “Level of significance”
Probability of Type I error – rejecting null hypothesis when it is in fact true
(false positive), typically 5%
p value = The probability of obtaining a result as extreme or more extreme than you found in your study by chance alone
Type I Error ( α) in Genetic and
Molecular Research
A genome-wide association scan of
500,000 SNPs will yield:
25,000 false positives by chance alone using
α = 0.05
5,000 false positives by chance alone using
α = 0.01
500 false positives by chance alone using
α = 0.001
Multiple Comparisons Problem
Multiple comparisons (or "multiple testing") problem occurs when one considers a set, or family, of statistical inferences simultaneously
Type I errors are more likely to occur
Several statistical techniques have been developed to attempt to adjust for multiple comparisons
Bonferroni adjustment
Adjusting alpha
Standard Bonferroni correction
Test each SNP at the α* =α /m
1 level
Where m
1
= number of markers tested
Assuming m
1
= 500,000, a Bonferroni-corrected threshold of α*= 0.05/500,000 = 1x10–7
Conservative when the tests are correlated
Permutation or simulation procedures may increase power by accounting for test correlation
Measures of LD
Jess Paulus, ScD
January 29, 2013
Haplotype definition
Haplotype: an ordered sequence of alleles at a subset of loci along a chromosome
Moving from examining single genetic markers to sets of markers
Measures of linkage disequilibrium
a g a g A G A G
A a
G g
A
A
G g
A
A g
G a
A
A G A G a g a
Basic data: table of haplotype frequencies
G g
A
8
2
62.5% a
0
6
37.5%
50%
50% g g
G
D’ and r
2
are most common
Both measure correlation between two loci
D prime …
Ranges from 0 [no LD] to 1 [complete LD]
R squared…
also ranges from 0 to 1
is correlation between alleles on the same chromosome
D
Deviation of the observed frequency of a haplotype from the expected is a quantity called the linkage disequilibrium (D)
If two alleles are in LD, it means D ≠ 0
If D=1, there is complete dependency between loci
Linkage equilibrium means D=0
Q
*
G g
Measure
D’
2 = r 2
A n
11 n
01 n
1 a n
10 n
00 n
0
Formula n
11 n
00
n
10 n
01 min( n
1 n
0
, n
0 n
1
)
n
11 n
00
n
10 n
01
2 n
1 n
0 n
1
n o
n
11 n
00
n
10 n
11 n
0 n
01 n
11 n
00 n n
11
11 n n n
10
00
00 n
01
n
10 n
01
n
10 n
01 n
1
n
0
Ref.
Lewontin (1964)
Hill and Weir
(1994)
Levin (1953)
Edwards (1963)
Yule (1900)
a
A a
A g
G g
G a
A
A
A g
G g
G
A
A
A a
G g
G g
A a
A a
G g
G g
D
’
= n
11 n
00
n
10 n
01 min( n
1 n
0
, n
0 n
1
)
A
G 8 g a
0
2 6
62.5% 37.5%
50%
50%
R 2 =
n
11 n
00
n
10 n
01
2 n
1 n
0 n
1
n o
D’ = (8
6 – 0x2) / (8
6) =1 r 2 = (8
6 – 0x2) 2 / (10
6
8
8)
= .6
r
2
and power
r 2 is directly related to study power
A low r 2 corresponds to a large sample size that is required to detect the LD between the markers
r 2 *N is the “effective sample size”
If a marker M and causal gene G are in LD, then a study with N cases and controls which measures M
(but not G) will have the same power to detect an association as a study with r 2 *N cases and controls that directly measured G
r
2
and power
Example:
N = 1000 (500 cases and 500 controls) r 2 = 0.4
If you had genotyped the causal gene directly, would only need a total N=400 (200 cases and
200 controls)
Today’s topics
1.
2.
Multiple comparisons
Measures of Linkage disequilibrium
• D’ and r 2
• r 2 and power