visualization - National Alliance for Medical Image Computing
advertisement
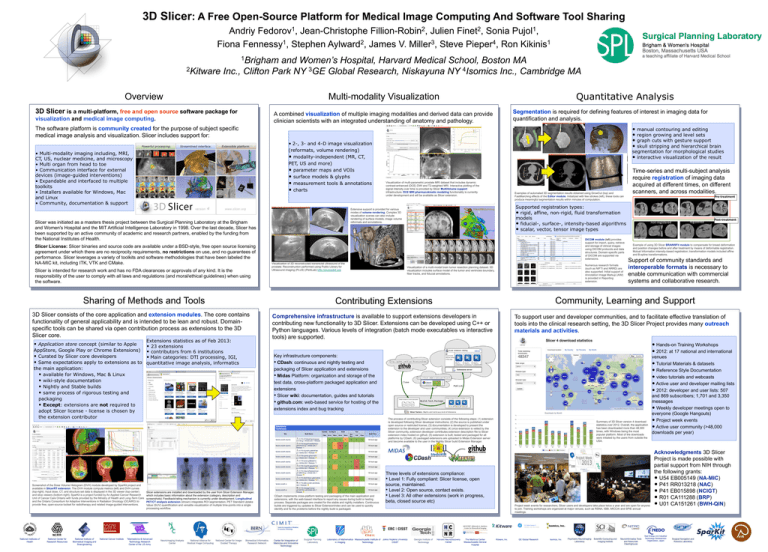
3D Slicer: A Free Open-Source Platform for Medical Image Computing And Software Tool Sharing 1 Fedorov , 2 Fillion-Robin , 2 Finet , 1 Pujol , Andriy Jean-Christophe Julien Sonia 1 2 3 4 1 Fiona Fennessy , Stephen Aylward , James V. Miller , Steve Pieper , Ron Kikinis 1Brigham and Women’s Hospital, Harvard Medical School, Boston MA 2Kitware Inc., Clifton Park NY 3GE Global Research, Niskayuna NY 4Isomics Inc., Cambridge MA Overview 3D Slicer is a multi-platform, free and open source software package for visualization and medical image computing. Quantitative Analysis Multi-modality Visualization A combined visualization of multiple imaging modalities and derived data can provide clinician scientists with an integrated understanding of anatomy and pathology. Segmentation is required for defining features of interest in imaging data for quantification and analysis. • manual contouring and editing • region growing and level sets • graph cuts with gesture support • skull stripping and hierarchical brain The software platform is community created for the purpose of subject specific medical image analysis and visualization. Slicer includes support for: • 2-, 3- and 4-D image visualization (reformats, volume rendering) • modality-independent (MR, CT, PET, US and more) • parameter maps and VOIs • surface models & glyphs • measurement tools & annotations • charts • Multi-modality imaging including, MRI, CT, US, nuclear medicine, and microscopy • Multi organ from head to toe • Communication interface for external devices (image-guided interventions) • Expandable and interfaced to multiple toolkits • Installers available for Windows, Mac and Linux • Community, documentation & support Slicer is intended for research work and has no FDA clearances or approvals of any kind. It is the responsibility of the user to comply with all laws and regulations (and moral/ethical guidelines) when using the software. Sharing of Methods and Tools 3D Slicer consists of the core application and extension modules. The core contains functionality of general applicability and is intended to be lean and robust. Domainspecific tools can be shared via open contribution process as extensions to the 3D Slicer core. Extensions statistics as of Feb 2013: • Application store concept (similar to Apple • 23 extensions AppStore, Google Play or Chrome Extensions) • contributors from 6 institutions • Curated by Slicer core developers • Main categories: DTI processing, IGI, • Same expectations apply to extensions as to quantitative image analysis, informatics the main application: • available for Windows, Mac & Linux • wiki-style documentation • Nightly and Stable builds • same process of rigorous testing and packaging • Except: extensions are not required to adopt Slicer license - license is chosen by the extension contributor Visualization of multi-parametric prostate MRI dataset that includes dynamic contrast-enhanced (DCE) DWI and T2-weighted MRI. Interactive plotting of the signal intensity over time is provided by Slicer MultiVolume support infrastructure. DCE MRI pharmacokinetic modeling functionality is currently under development and will be available as Slicer extension. Examples of automated 3D segmentation results obtained using GrowCut (top) and FastMarching effects of the Editor module. Initialized with few strokes (left), these tools can produce meaningful segmentation results within minutes of computation. DICOM module (left) provides support for import, query, retrieve and storage of clinical images using DICOM protocols and data structures. Domain-specific parts of DICOM are supported via extensions. Visualization of 3D reconstructed transrectal ultrasound of the prostate. Reconstruction performed using Public Library for Ultrasound imaging (PLUS) (PerkLab) http://plustoolkit.org Time-series and multi-subject analysis require registration of imaging data acquired at different times, on different scanners, and across modalities. Pre-treatment Supported registration types: • rigid, affine, non-rigid, fluid transformation models • fiducial-, surface-, intensity-based algorithms • scalar, vector, tensor image types Extensive support is provided for various modes of volume rendering. Complex 3D visualization scenes can also include rendering of surface models, image volume reformats and annotations. Slicer was initiated as a masters thesis project between the Surgical Planning Laboratory at the Brigham and Women's Hospital and the MIT Artificial Intelligence Laboratory in 1998. Over the last decade, Slicer has been supported by an active community of academic and research partners, enabled by the funding from the National Institutes of Health. Slicer License: Slicer binaries and source code are available under a BSD-style, free open source licensing agreement under which there are no reciprocity requirements, no restrictions on use, and no guarantees of performance. Slicer leverages a variety of toolkits and software methodologies that have been labeled the NA-MIC kit, including ITK, VTK and CMake. segmentation for morphological studies • interactive visualization of the result Visualization of a multi-modal brain tumor resection planning dataset. 3D visualization includes surface model of the tumor and ventricles boundary, fiber tracks, and fiducial annotations. Numerous research formats (such as NIFTI and NRRD) are also supported. Initial support of Annotation Image Markup (AIM) is provided in Reporting extension. non-registered Support of community standards and interoperable formats is necessary to enable communication with commercial systems and collaborative research. Community, Learning and Support Contributing Extensions Comprehensive infrastructure is available to support extensions developers in contributing new functionality to 3D Slicer. Extensions can be developed using C++ or Python languages. Various levels of integration (batch mode executables vs interactive tools) are supported. To support user and developer communities, and to facilitate effective translation of tools into the clinical research setting, the 3D Slicer Project provides many outreach materials and activities. • Hands-on Training Workshops • 2012: at 17 national and international Key infrastructure components: • CDash: continuous and nightly testing and packaging of Slicer application and extensions • Midas Platform: organization and storage of the test data, cross-platform packaged application and extensions • Slicer wiki: documentation, guides and tutorials • github.com: web-based service for hosting of the extensions index and bug tracking A B The process of contributing Slicer extension consists of the following steps: (1) extension is developed following Slicer developer instructions; (2) the source is published under open source or restricted license; (3) documentation is developed to present the extension to the developer and user communities; (4) once extension is vetted by the Slicer community, extension developer contributes extension description file to Slicer extension index hosted on github; (5) extension is built, tested and packaged for all platforms by CDash; (6) packaged extensions are uploaded to Midas Extension server and become available to the user in the Nightly Slicer build Extension Manager. Slicer extensions are installed and downloaded by the user from Slicer Extension Manager, which includes basic information about the extension (category, description and screenshots). Feedback/rating mechanism is currently under development. Longitudinal PET/CT analysis extension (shown) integrates ROI segmentation, PET Standard Uptake Value (SUV) quantification and versatile visualization of multiple time-points into a single processing workflow. CDash implements cross-platform testing and packaging of the main application and extensions, with the web-based interface to report any issues during build or testing process. Separate packages are created for the stable and nightly installers. Continuous builds are triggered by updates to Slicer ExtensionIndex and can be used to quickly identify and fix the problems before the nightly build is packaged. Post-treatment Example of using 3D Slicer BRAINSFit module to compensate for breast deformation and position changes before and after treatment by means of deformable registration. Mutual Information intensity-based registration; transformation models included affine and B-spline transformations. Summary of 3D Slicer version 4 download statistics over 2012. Overall, the application has been downloaded more than 48,000 times, with Windows being the most popular platform. Most of the downloads were initiated by the users from outside the USA. D Screenshot of the Dose Volume Histogram (DVH) module developed by SparKit project and available in SlicerRT extension. The DVH module compute metrics (left) and DVH curves (top-right). Input dose, CT, and structure set data is displayed in the 3D viewer (top-center) and slice viewers (bottom-right). SparKit is a project funded by An Applied Cancer Research Unit of Cancer Care Ontario with funds provided by the Ministry of Health and Long-Term Care and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions. registered Three levels of extensions compliance: • Level 1: Fully compliant: Slicer license, open source, maintained. • Level 2: Open source, contact exists. • Level 3: All other extensions (work in progress, beta, closed source etc) Project week events for researchers, Slicer users and developers take place twice a year and are open for anyone to join. Training workshops are organized at major venues, such as RSNA, ISBI, MICCAI and SPIE annual meetings. venues • Tutorial Materials & datasets • Reference Style Documentation • video tutorials and webcasts • Active user and developer mailing lists • 2012: developer and user lists: 507 and 869 subscribers; 1,701 and 3,350 messages • Weekly developer meetings open to everyone (Google Hangouts) • Project week events • Active user community (>48,000 downloads per year) Acknowledgments 3D Slicer Project is made possible with partial support from NIH through the following grants: • U54 EB005149 (NA-MIC) • P41 RR013218 (NAC) • P41 EB015898 (NCIGT) • R01 CA111288 (BRP) • U01 CA151261 (BWH-QIN)