Poster - Hughes Undergraduate Biology
advertisement

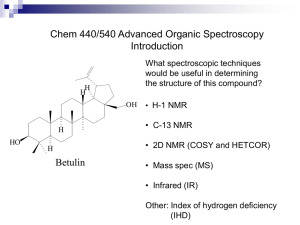
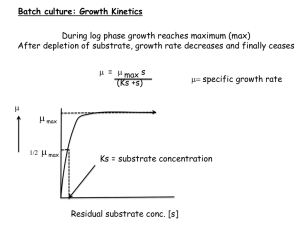
April, 2011 NMR Metabolite Profiling of Bacterial Biofilms in Chronic Wounds K.L. Morrissey, L.K. Jennings, P.R. Secor, B. Tripet, G.A. James, P.S. Stewart, V. Copié Department of Chemistry and Biochemistry, MSU’s NMR and MS Metabolomics Research Facilities METHODS PRELIMINARY RESULTS We hypothesize that anaerobic bacterial metabolism occurs deep within the wound bed where oxygen concentrations are low, and that the presence of fermenting microorganisms can be detected through NMR metabolite profiling. However, if adequate blood circulation is present near the bottom of the wound bed, anaerobic metabolism may be more active in the middle of the biofilm. PRELIMINARY RESULTS Intensity lactate acetate BCM - biofilm PCM - planktonic EPI – media control Sum line glucose Intensity histamine hydroxyphenylacetate histamine Chemical Shift (ppm, upfield) formate Chronic-wound biofilms are host-pathogen environments that harbor a large diversity of bacterial species including anaerobic, aerobic, and facultative organisms. Metabolomic profiling of chronic wounds may provide a more accurate indication of the average oxidative state of cells within the biofilm allowing treatment to be tailored to individual patients. Metabolomics is an emerging field that involves the quantification of small-molecule metabolites (<1000 Da) in bodily fluids, tissues, or cells to gain insight into the operation of biological systems. glutamine Chronic wounds including diabetic foot ulcers, venous leg ulcers, and pressure ulcers are healing-resistant wounds that are characterized by prolonged inflammation and failure to re-epithelialize. The presence of a biofilm is increasingly accepted as the major impediment to wound healing. Disruption of the biofilm community and removal of necrotic tissue via debridement has proven successful in the treatment of chronic wounds. Unfortunately, it is difficult to completely remove the biofilm, and incomplete removal can result in inability to clear the infection and, ultimately, limb amputations. glutamine INTRODUCTION Figure 2. Bruker 600 equipped with sample jet Cell metabolites were extracted using a methanol/chloroform/water extraction method. 1-D NOESY 1H NMR spectrum were acquired using a Bruker 600 MHz NMR equipped with an autosampler. The NMR spectra were processed and the metabolites quantified using Chenomx software suite. Quantification of the metabolites were determined by comparison to the internal standard 4,4-dimethyl-4silapentane-1-sulfonic acid (DSS). Hierarchical clustering conducted in Genesis was used to visualize results. Peg Dirckx CBE Figure 1. Illustration of chronic wound micro-community where biofilms are a major barrier to wound healing. OBJECTIVE The goal of this study is to use NMR metabolite profiling to analyze the extracellular metabolome of Staphylococcus aureus biofilms compared to planktonically grown cells. S. aureus is an important human pathogen and a predominant organism found in chronic-wound microcommunities. Metabolite profiling of S. aureus excretions may provide insight into the mechanism of pathogenesis and the persistence of infection in chronic wounds. 7 7 Wound tissue metabolites were extracted also using a methanol/chloroform/water extraction method. Similar to the method with the cells, 1-D NOESY 1H NMR spectrum were acquired and the NMR spectra were processed and the metabolites quantified using Chenomx software suite. Tissue Sample Conc. ( mM ) Metabolite Serum Sample Conc. ( mM ) Lactate 0.3866 Glucose 6.4612 Glucose 0.3553 Lactate 2.5069 Taurine 0.2008 Glutamine 0.9771 Fructose 0.1195 Alanine 0.7244 Homoserine 0.1058 Glycerol 0.4299 Alanine O-Phosphoethanolamine Glycerol 0.0755 Valine 0.3507 0.0703 Threonate 0.2725 0.066 Propylene glycol 0.2392 Glutamate 0.0617 Lysine 0.2247 Glutamine 0.0614 Citrate 0.2218 Table 1. Most abundant metabolite concentrations found in the right foot medial tissue and corresponding serum sample in the NMR tube. This data must be normalized in the future for further analysis and comparison. Note: Further validation of metabolites' presence and concentration using 2D NMR and mass spectrometry is in progress. CONCLUSIONS Chemical Shift (ppm, downfield) Figure 3. NMR spectra of excreted metabolites from S. aureus biofilms (blue), planktonic cells (green), and EpiLife growth medium (EPI, black). PCM BCM EPI Preliminary results indicate that glucose and amino acids were selectively consumed by S. aureus biofilms, while mixed-acid fermentation products (lactate, acetate, and formate) were produced. The results agree with previously published findings from proteomics, transcriptomics, and identification of individual metabolite studies that suggest that anaerobic or microaerobic metabolism is important to the S. aureus biofilm phenotype. NMR-metabolite profiling was effective at quantifying metabolites in S. aureus biofilm and planktonic cells. Metabolite profiling of S. aureus excretions revealed metabolites unique to biofilms and may provide insight into the mechanism of pathogenesis and the persistence of infection in chronic wounds. Pressure ulcer Samples: Pressure ulcer biopsies Serum samples Metabolite Low Conc. High Conc. Mixed-acid fermentation products formed by biofilm Preliminary results of the chronic wound and serum profiling demonstrate abundant metabolites found in the corresponding samples in the NMR tube. The serum control can help delineate host metabolites from biofilm metabolites once the data can be accurately compared. ACKNOWLEDGMENTS Amino acids in media selectively consumed by biofilm and planktonic cells Serum metabolites were extracted using a 3 kDa Amicon Ultra 0.5 ml filter. The filter was washed 5 times with 0.5 ml of sterile water and centrifuged for 12 minutes at 14,000 g. The Filter is coated with a small molecule membrane preservative, such as glycerol, which can interfere with metabolite analysis. Adding these rinsing steps can improve the recovery of metabolites from the protein component. Following the washing, 1 ml of serum, split between two filters (0.5 ml in each filter), was filtered through the washed filter by centrifuging for 30 minutes at 14,000 g. Figure 4. Hierarchical clustering (covariance distance) of S. aureus metabolites from triplicate PCM, BCM, and EPI. Light colors indicate high metabolite concentrations, while black indicates metabolite concentrations below detection. The serum filtrate was combined with 10% D2O NMR buffer and 1-D NOESY 1H NMR spectrum were acquired. Metabolites were again quantified using Chenomx software suite. Note: Further validation of metabolites' presence and concentration using 2D NMR and mass spectrometry is in progress. C-source Funding was provided by the Undergraduate Hughes Scholars Program through the Howard Hughes Medical Institute (Grant #52006931) and NCRR Administrative Supplement Award to “Advance T1 & T2 Translational Research” to NIH P20 RR024237 COBRE Award. The NMR Metabolomics Facility at MSU is supported by an Administrative Supplement Award to Dr. Dratz “Center for the Analysis of Cellular Mechanisms and Systems Biology”. We thank MSU COBRE director, Dr. Ed Dratz, as well as Dr. Brian Bothner, and Dr. Jonathan Hilmer, Director and Manager, respectively, of MSU’s Mass Spectrometry and Proteomics Facility. We also thank the Medical Biofilms Laboratory at the Center for Biofilm Engineering, Al Parker, Scott Busse, and Kate McInnerney for technical assistance.