10/31
advertisement
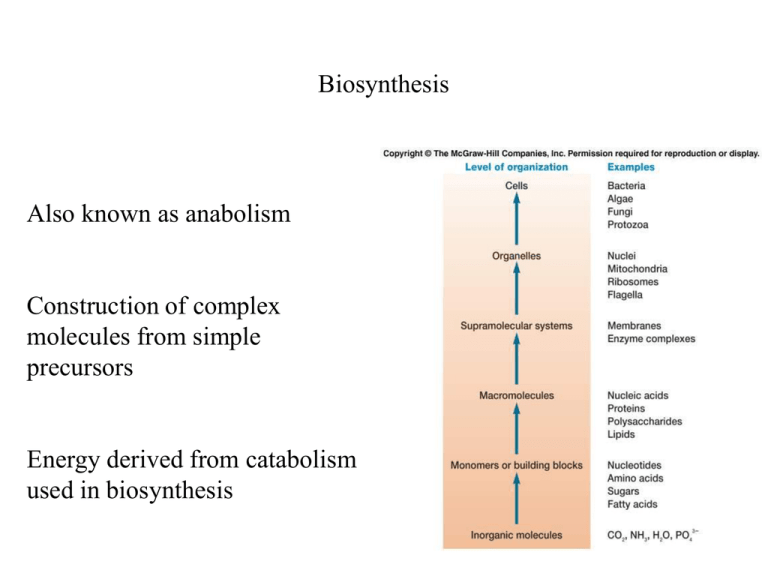
Biosynthesis Also known as anabolism Construction of complex molecules from simple precursors Energy derived from catabolism used in biosynthesis Sulfur assimilation Sulfur is required for the formation of cysteine, methionine and many cofactors Sulfate (SO42) is often used as a source of sulfur Sulfate must be reduced before assimilation assimilatory sulfate reduction Sulfur assimilation Sulfate is activated by the formation of phosphoadenosine5-phosphosulfate Sulfate is then reduced to sulfite (SO32) then to hydrogen sulfide (H2S) Sulfur assimilation Cysteine is then formed from H2S and used in the formation of other sulfur containing molecules Nitrogen assimilation Nitrogen required for proteins, nucleic acids and other important cell constituents Most microorganisms are incapable of using nitrogen gas as a nitrogen source They must therefore incorporate either ammonia (NH3) or nitrate (NO3) Ammonia incorporation Ammonia is easily incorporated because it is more highly reduced than other forms of nitrogen Can be combined with pyruvate to form alanine or ketoglutarate to form glutamate Ammonia incorporation Ammonia can also be incorporated using two enzymes acting in sequence Glutamine synthetase and glutamate synthetase Ammonia incorporation Ammonia used to synthesize glutamine from glutamate Amino group of glutamine transferred to -ketoglutarate to form 2 molecules of glutamate Amino group can then be transferred to form other amino acids Assimilatory nitrate reduction Nitrate must be converted to ammonia before incorporation into organic compounds Nitrate is first reduced to nitrite by nitrate reductase Assimilatory nitrate reduction Nitrite is reduced to ammonia by nitrite reductase Ammonia is then incorporated into organic material Nitrogen fixation The reduction of gaseous nitrogen to ammonia Rate of this process often limits plant growth Carried out by a small number of microorganisms Nitrogen fixation Reduction of nitrogen to ammonia is catalyzed by nitrogenase Sequential addition of electron pairs results in formation of 2 molecules of ammonia from 1 molecule of N2 Nitrogen fixation Energetically expensive: requires 8 electrons and 16 ATPs Synthesis of amino acids Carbon skeletons derived from acetyl-CoA and intermediates of glycolysis, the TCA cycle and the pentose phosphate pathway Synthesis of amino acids Synthesis of amino acids Common intermediates are used to synthesize families of related amino acids Synthesis of amino acids Common intermediates are used to synthesize families of related amino acids Anapleurotic reactions TCA cycle intermediates used for biosynthesis could be depleted Anapleurotic reactions serve to replenish cycle intermediates Anapleurotic reactions Most microorganisms replace TCA cycle intermediates by CO2 fixation Different from autotrophs since only used to replace intermediates Pyruvate or PEP used as acceptor molecule to form oxaloacetate Glyoxylate pathway Some microorganisms can use acetate as the sole carbon source Synthesize TCA cycle intermediates using the glyoxylate pathway Modified TCA cycle Glyoxylate pathway Isocitrate converted to succinate and glyoxylate Glyoxylate combines with acetyl-CoA to form oxaloacetate Prevents loss of acetyl-CoA carbons as CO2 Synthesis of purines and pyrimidines Cyclic nitrogen containing bases that are used in the synthesis of ATP, DNA, RNA and other cell components Purines contain two joined rings: adenine and guanine Pyrimidines have a single ring: cytosine, thymine and uracil Synthesis of purines and pyrimidines Purine or pyrimidine joined to pentose sugar (ribose or deoxyribose) = nucleoside Nucleoside + one or more phosphate group = nucleotide Synthesis of purines Seven different molecules contribute parts to final skeleton Synthesis of purines Inosinic acid is the first common intermediate Adenosine and guanosine monophosphates formed Nucleoside diphosphates and triphosphates formed by phosphate transfers from ATP Synthesis of pyrimidines Aspartic acid and carbamoyl phosphate combine Eventually converted to orotic acid Ribose then added and decarboxylation results in uridine monophosphate Synthesis of fatty acids Uses acetyl-CoA and malonylCoA as substrates Malonyl-CoA formed from acetyl-CoA and CO2 Both are transferred to acyl carrier protein (ACP) Synthesis of fatty acids Malonyl-ACP reacts with fatty acyl-ACP to yield CO2 and fatty acyl-ACP + 2 carbons Followed by 2 reductions and a dehydration Fatty acyl-ACP then ready to accept another malonyl-ACP Synthesis of fatty acids vs. -oxidation Reverse process except uses CoA as carrier rather than ACP Synthesis of lipids Dihydroxyacetone phosphate reduced to glycerol 3-P Glycerol 3-P combines with 2 fatty acids to form phosphatidic acid Attachment of third fatty acid yields triglyceride Synthesis of lipids Phosphatidic acid attached to cytidine diphosphate (carrier) Reacts with serine to form phosphatidylserine Decarboxylation yields phosphatidylethanolamine