Enzymes
advertisement

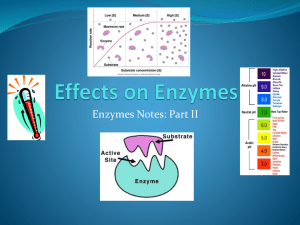
Enzymes and heart attacks Enzymes: “Helper” Protein molecules 2009-2010 Flow of energy through life Life is built on chemical reactions Chemical reactions of life Processes of life • building molecules • synthesis + • breaking down molecules • digestion + Nothing works without enzymes! How important are enzymes? • all chemical reactions in living organisms require enzymes to work • building molecules enzyme • synthesis enzymes + • breaking down molecules • digestive enzymes We can’t live without enzymes! • enzymes speed up reactions • “catalysts” enzyme + Examples synthesis + enzyme digestion enzyme + Enzymes are proteins Each enzyme is the specific helper to a specific reaction • each enzyme needs to be the right shape for the job • enzymes are named for the reaction they help • sucrase breaks down sucrose • proteases breakdown proteins Oh, I get it! • lipases breakdown lipids They end in -ase • DNA polymerase builds DNA Enzymes aren’t used up Enzymes are not changed by the reaction • used only temporarily • re-used again for the same reaction with other molecules • very little enzyme needed to help in many reactions substrate active site product enzyme It’s shape that matters! Lock & Key model • shape of protein allows enzyme & substrate to fit • specific enzyme for each specific reaction 2 1 3 Enzyme vocabulary Enzyme • helper protein molecule Substrate • molecule that enzymes work on Products • what the enzyme helps produce from the reaction Active site • part of enzyme that substrate molecule fits into What affects enzyme action Correct protein structure • correct order of amino acids • why? enzyme has to be right shape Temperature • why? enzyme has to be right shape pH (acids & bases) • why? enzyme has to be right shape Order of amino acids Wrong order = wrong shape = can’t do its job! chain of amino acids DNA folded protein right shape! folded protein chain of amino acids DNA wrong shape! Temperature Effect on rates of enzyme activity • Optimum temperature • greatest number of collisions between enzyme & substrate • human enzymes • 35°- 40°C (body temp = 37°C) • Raise temperature (boiling) • denature protein = unfold = lose shape • Lower temperature T° • molecules move slower • fewer collisions between enzyme & substrate Temperature reaction rate human enzymes 37° temperature What’s happening here?! How do cold-blooded creatures do it? pH Effect on rates of enzyme activity • changes in pH changes protein shape • most human enzymes = pH 6-8 • depends on where in body • pepsin (stomach) = pH 3 • trypsin (small intestines) = pH 8 pH intestines trypsin What’s happening here?! reaction rate stomach pepsin 0 1 2 3 4 5 6 pH 7 8 9 10 11 12 13 14 For enzymes… What matters? SHAPE! 2009-2010 Let’s build some Enzyme Models! 2009-2010 Myocardial infarction Acute myocardial infarction is the rapid development of myocardial necrosis caused by a critical imbalance between the oxygen supply and demand of the myocardium. 500,000-700,000 deaths in the US annually. Myocardial infarction Symptoms • Angina pectoralis • Dyspnea • Nausea and/or abdominal pain • Anxiety • Lightheadedness and syncope • Cough • Nausea and vomiting • Diaphoresis One problem Differential diagnosis • Pericarditis • Aortic Dissection • Cholecystitis and Cholelithiasis • Laryngeal spasm • Anxiety attack • and on and on and on… One solution – “Cardiac enzymes” Enzymes Definition: Biological catalysis Qualities Fe3 2 H 2O2 2 H 2O O2 • Efficient catalase • Specific • Stereo-specific - they can tell the difference between isomers • Regulated • Saturable • Inhibitable substrate enzyme product (s) Substrate versus product Types of enzymes All enzymes end in the suffix “_______ase” Different versions of the same enzyme (often made by alternative splicing) are called isoenzymes or isozymes General classes of enzymes • • • • • • Polymerases – nucleic acid synthesis Transferases – transfer a functional group Hydrolases – hydrolytic cleavage Proteases – hydrolytic cleavage of protein chains Kinases – add phosphate groups to compounds … and many, many more… Mechanism Enzymes work by lowering activation energy • If you don’t understand free energy changes, see Box 5A in your book ∆G is a measure of the ability of a reaction to go forward, but not necessarily the rate EA is the activation energy. The rate at which a reaction proceeds is directly proportional to the number of molecules reaching the transition state - that is, those that reach EA. Things for optimal activity pH – alters enzyme structure by altering charge Temperature – increases activity by moving molecules closer to the activation energy, and by making ∆G slightly more negative… until the enzyme "denatures" Coenzymes – like biotin in amino group transfer – bind reversibly but participate directly Metal ions – like magnesium in some ATPases. Michaelis-Menten Kinetics Shows saturation at high substrate concentrations Vmax – rate at saturation for a given enzyme concentration in moles per unit time Km – Michaelis constant – substrate concentration that gives ½ maximal velocity Vmax S V K m S How do you measure this crap? Things you need: • The enzyme • The substrate • A way of measuring either the disappearance of substrate, or the appearance of product, usually photometrically. Other commonly reported values Turnover • rate at saturation for 1 enzyme molecule (reactions catalyzed per second per molecule) “Units” • are defined by convention, but are something of an industry standard. For example… • “One unit of creatine kinase is defined as the amount necessary to catalyze the conversion of one micromole of creatine to creatine phosphate per minute at 25°C and pH 8.9.” Competitive inhibitors Many drugs (like Cipro and anti-HIV drugs) are enzyme inhibitors Two major kinds of inhibitors: competitive and noncompetitive. Competitive inhibitors bind to the active site of the enzyme. Alter Km but not Vmax. What will happen to V if you push the substrate concentration very high? Noncompetitive inhibitors Noncompetitive inhibitors bind somewhere besides the active site. They alter the behavior of the enzyme in a manner analogous to allosteric regulation Alter Vmax. What will happen to V if you push the substrate concentration very high? Regulation Allosteric regulation A regulatory molecule binds to a site separate from the active site (like small molecules to repressors in operons) Induced conformational changes regulate the activity of the enzyme These enzymes usually have catalytic and regulatory domains Can have multiple domains or subunits for different regulators Regulation Allosteric Cooperativity • One substrate aids or impedes the catalysis of another • Implies multiple catalytic subunits. Covalent modification • Adding/removing groups – like phosphate groups by kinases • Cleaving bonds – converting proenzymes to enzymes like in the blood clotting cascade Association-dissociation of subunits • One protein binds to another, thereby activating the enzymatic activity of one of them. Creatine kinase Creatine phosphate acts as a backup for rapid ATP regeneration in active tissues • Creatine phosphate is in energetic equilibrium with ATP • Creatine kinase (CK) catalyzes the transfer of phosphate between creatine and ATP/ADP Provides rapid regeneration of ATP when ATP is low Creatine phosphate is regenerated when ATP is abundant ADP ATP CK Cr-P Cr Application: Cardiac enzymes enzymes released from injured myocardium. Creatine kinase (CK) is the one usually assayed If CK is found in the blood stream, this implies that the myocardium may have been damaged Problems: • Tells you little about the time course or severity • Lets you spot really small infarcts. • What else? Creatine kinase isozymes The enzyme is dimeric Two different polypeptide chains (M and B) are differentially expressed in tissues Combine at random to give three isozymes: • CK-MM (primarily muscle) • CK-MB (hybrid) • CK-BB (primarily brain) The CK-MB has its highest concentration in heart muscle CK-MB >5% of total CPK strongly suggests myocardial infarction Determining CK-MB (mass) / CK (activity) Total CK activity is determined by a simple enzyme assay (phosphocreatine + ADP ATP) CK-MB mass is determined by a two-antibody “sandwich” assay. Y Y Y Y Y Y Y Y Y Y Y Y Y Y Y Y Substrate Y Y Y Y Y Tagged anti-CK-M Y Y Y Y Y anti-CK-B coated tube Y Y Y Y Y Y Y Y Y Y Y Y Y Y Y Y Y Y POSITIVE