Lecture 25
advertisement

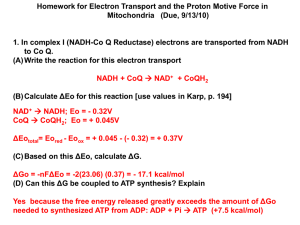
Lecture 25 The sources of energy: posphorylation, oxidation and coupling chemical energy to work Peter Mitchell Why protons? Why ATP? Why oxygen? Most cells use a proton gradient as an energy source across their plasma membrane. Why do animal cells use a sodium gradient? Why protons? Protons because one single Histidine, Glutamate or Aspartate residue furnish a simple and tunable binding site for H+. pKa of these groups can vary by 2-3 units depending on the environment. These sites do not bind metal ions tightly unless work in concert. It is much more difficult to build a selective binding site that would discriminate between Na+, K+, Mg2+, Ca2+, Zn2+, Cu2+, Pb2+, Hg2+, Fe2+, Fe3+, etc Protomotive Force has an electrical component and can couple electrochemical proton gradient to the transport of other charged substances H RT ln( H [ H ]1 ) F ... in Joules [ H ]2 60 pH ..... in mV F 0 mV -180 mV pH = 8 PMF = -60 -180 = -240 mV pH = 7 Measuring the membrane potential….. 0 mV -180 mV positive charge pH = 7 pH = 8 Calibration??? Rhodamine 123 Fluorescent dye Rhodamine 123 (Rh123+) will penetrate into the vesicles according to electric gradient, increasing their fluorescence. Increased concentration of Rh123 inside the vesicle beyond certain point will cause self-quenching. Measuring the membrane potential…..another way 0 mV -180 mV pH = 7 pH = 8 K+ Valinomycin…potassium uniporter What happens if we add valinomycin? RT F ln( [ K ] out ) [ K ]in but val. may affect the potential… http://www.rpi.edu/dept/bcbp/molbiochem/MBWeb/mb1/part2/carriers.htm Measuring the membrane potential…..a better way val [KCl] outside mM fluorescence 0.05 0.1 0.2 time Null method Why ATP? phosphoanhydride bonds The reaction of ATP hydrolysis is very favorable ΔGo = -30.5 kJ/mol = - 7.3 kCal/mol because: 1. Charge separation of closely packed phosphate groups provides electrostatic relief 2. Inorganic Pi, the product of the reaction, is immediately resonance-stabilized (electron density spreads equally to all oxygens) 3. ADP immediately ionizes giving H+ into a low [H+] environment (pH~7) 4. Both Pi and ADP are more favorably solvated by water than one ATP molecule. Mg2+ ATP exists in complex with Mg2+ ATP is not the only: Phosphoenolpyruvate (PEP) -61.9 kJ/mol 1,3-Bisphosphoglycerate -49.3 kJ/mol Phosphocreatinine -43.0 kJ/mol ATP -30.5 kJ/mol Pyrophisphate (Pi-Pi) or Inorganic polyphosphate (polyPi) -19 kJ/mol Thioesters (Acetyl CoA) -31 kJ/mol In real cells G for ATP hydrolysis is more negative than standard Go G p G o RT ln [ADP][P] [AT P] Calculate Gp in erythrocytes if Gp = -30.5 kJ/mol [ATP] = 2.3 mM; [ADP] = 0.25; [Pi] = 1.65 mM, In other cells: Rat myocyte Rat neuron E. coli [ATP] 8 2.6 7.9 [ADP] [AMP] 0.9 0.04 0.73 0.06 1.04 0.82 [Pi] in mM 8.05 2.7 7.9 ATP provides energy to group transfer reactions: A-P → A + P G1 B-P → B + P G2 A-P + B → A + B-P G = G1-G2 G of hydrolysis PEP 1,3-BPG P E P Phosphocreatinie ATP Glucose-6-P Pi Glycerol-P Synthesis of any phosphorylated compound can be coupled to ATP hydrolysis Transfer reactions: Phoshoryl transfer; Pyrophosphoryl transfer; Adenylyl transfer Synthesis of NTPs (dNTPs) from ATP occurs as phosphoryl exchange at G ~0. The reaction is catalyzed by nucleoside diphosphate kinase which first phosphorylates its own His, releases ADP and then phosporylates the incoming NDP or dNDP. How is the energy of phosphoryl transfer (or removal) imparted to a conformational change or how the chemical work is done? Simple binding of ATP to an enzyme (or another effector protein) may cause massive conformational change by allosteric mechanism through the stage of binding site rearrangement. Cleavage of the gamma phosphate would lead to another conformational change. A complete release of the nucleotide diphosphate returns the protein to its initial state. Phosphorylation of certain sites (Tyr, Ser or Thr) promotes recognition by a counterpart domain (recall SH2 domains) Why oxygen? Electron re-distribution from less electronegative to more electronegative atoms occurs with massive energy release: Electronegativity of common elements: H < C < S < N < O …Cl < F Identify the substance and the reduction state of the first carbon (red) Enthalpies of oxidation (combustion) hydrogen (MW 2) H2 + ½O2 → H2O -286 kJ/mol methane (MW 16) CH4 + 3O2 → CO2 + 2H2O -891 kJ/mol glucose (MW 180.2) C6H12O6 + 6O2 → 6CO2 + 6H2O palmitic acid (MW 256.4) C16H32O2 + 23O2 – 16CO2 + 16H2O -2840 kJ/mol (-680 kCal/mol) - 9730 kJ/mol Oxidation-reduction (Red-ox) reactions usually lead to redistribution of electron densities or complete transfer of electrons resulting in change of ionization state. Fe2+ + Cu2+ → Fe3+ + Cu+ or in the form of half-reactions: Fe2+ → Fe3+ + e Cu2+ + e → Cu+ In biological systems oxidation is often coupled to dehydrogenation. 1. Direct transfer of electrons 2. As a transfer of H atoms or removal of H atoms coupled to production of H+ 3. As a Hydride ion :H– 4. Through direct combination with oxygen R-CH3 + (½)O2 → R-CH2-OH Standard Reduction potentials for some half-reactions, Volt ½ O2 + 2H+ + 2e → H2O +0.816 Fe3+ + e → Fe2+ +0.771 Cytochrome c (Fe3+) + e → Cytochrome c (Fe2+) +0.254 Fumarate2- +2H+ + 2e → succinate2- +0.031 2H+ + 2e → H2 0 (standard condition) Pyruvate + 2H+ + 2e → lactate -0.185 FAD + 2H+ + 2e → FADH2 -0.219 S + 2H+ + 2e → H2S -0.243 NAD+ + H+ + 2e → NADH -0.320 NADP+ + H+ + 2e → NADPH -0.324 α-ketoglutarate + CO2 + 2H+ + 2e → isocytrate -0.38 2H+ + 2e → H2 -0.414 (pH 7) Reduction potentials for mixtures of reductant/oxidant (hlfreaction potentials) are measured using the standard hydrogen electrode RT [electron acceptor] o E E ln nF [electron donor] 2H+ + 2e → H2 Mg2+ + 2e → Mg (metal) http://www.chemguide.co.uk/physical/redoxeqia/eomgdiag.gif Electron transport chain puts reductants and oxidants in specific order Complex I II III IV Energy released in forming water is stored as a PMF Energy is divided into smaller units ~12 protons per water molecule