ICBICposter05sized
advertisement
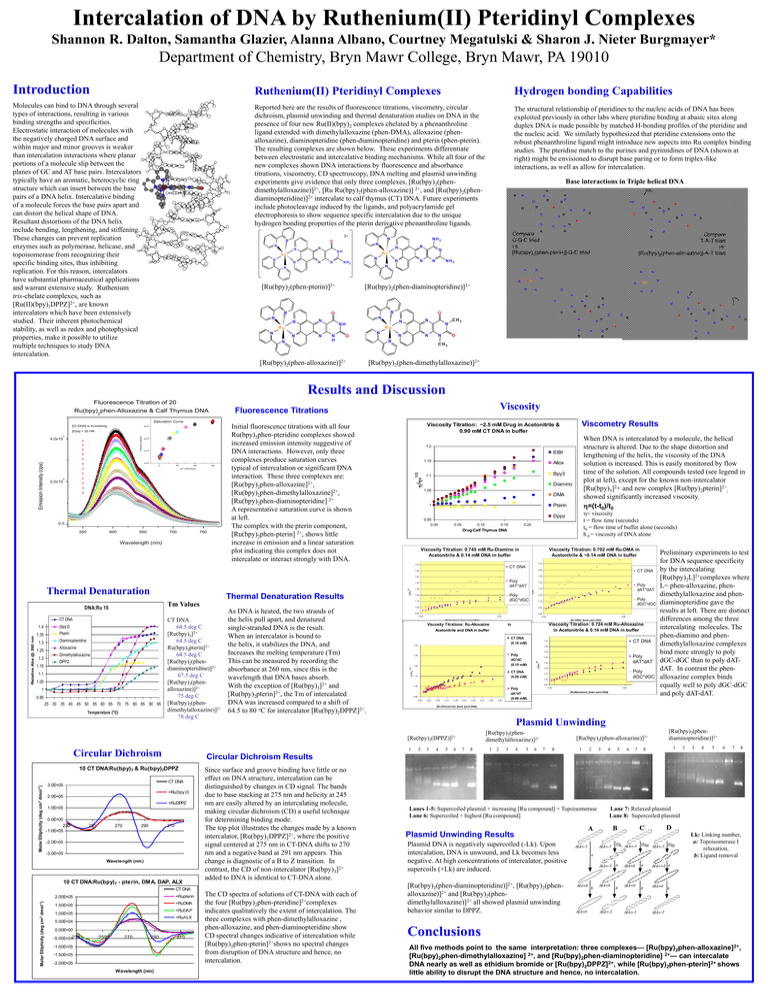
Intercalation of DNA by Ruthenium(II) Pteridinyl Complexes Shannon R. Dalton, Samantha Glazier, Alanna Albano, Courtney Megatulski & Sharon J. Nieter Burgmayer* Department of Chemistry, Bryn Mawr College, Bryn Mawr, PA 19010 Introduction Ruthenium(II) Pteridinyl Complexes Hydrogen bonding Capabilities Molecules can bind to DNA through several types of interactions, resulting in various binding strengths and specificities. Electrostatic interaction of molecules with the negatively charged DNA surface and within major and minor grooves is weaker than intercalation interactions where planar portions of a molecule slip between the planes of GC and AT base pairs. Intercalators typically have an aromatic, heterocyclic ring structure which can insert between the base pairs of a DNA helix. Intercalative binding of a molecule forces the base pairs apart and can distort the helical shape of DNA. Resultant distortions of the DNA helix include bending, lengthening, and stiffening. These changes can prevent replication enzymes such as polymerase, helicase, and topoisomerase from recognizing their specific binding sites, thus inhibiting replication. For this reason, intercalators have substantial pharmaceutical applications and warrant extensive study. Ruthenium tris-chelate complexes, such as [Ru(II)(bpy)2DPPZ]2+, are known intercalators which have been extensively studied. Their inherent photochemical stability, as well as redox and photophysical properties, make it possible to utilize multiple techniques to study DNA intercalation. Reported here are the results of fluorescence titrations, viscometry, circular dichroism, plasmid unwinding and thermal denaturation studies on DNA in the presence of four new Ru(II)(bpy)2 complexes chelated by a phenanthroline ligand extended with dimethylalloxazine (phen-DMA), alloxazine (phenalloxazine), diaminopteridine (phen-diaminopteridine) and pterin (phen-pterin). The resulting complexes are shown below. These experiments differentiate between electrostatic and intercalative binding mechanisms. While all four of the new complexes shown DNA interactions by fluorescence and absorbance titrations, viscometry, CD spectroscopy, DNA melting and plasmid unwinding experiments give evidence that only three complexes, [Ru(bpy)2(phendimethylalloxazine)]2+, [Ru Ru(bpy)2(phen-alloxazine)] 2+, and [Ru(bpy)2(phendiaminopteridine)]2+ intercalate to calf thymus (CT) DNA. Future experiments include photocleavage induced by the ligands, and polyacrylamide gel electrophoresis to show sequence specific intercalation due to the unique hydrogen bonding properties of the pterin derivative phenanthroline ligands. The structural relationship of pteridines to the nucleic acids of DNA has been exploited previously in other labs where pteridine binding at abasic sites along duplex DNA is made possible by matched H-bonding profiles of the pteridine and the nucleic acid. We similarly hypothesized that pteridine extensions onto the robust phenanthroline ligand might introduce new aspects into Ru complex binding studies. The pteridine match to the purines and pyrimidines of DNA (shown at right) might be envisioned to disrupt base paring or to form triplex-like interactions, as well as allow for intercalation. Base interactions in Triple helical DNA C H3 H R N H N H N N O N N N N N NH N N N N N N N N H2 N N N N R H N N N [Ru(bpy)2(phen-diaminopteridine)]2+ N O N N N NH N N H H N N R N O N H N Compare T-A-T triad vs. [Ru(bpy)2(phen-alloxazine)]-A-T triad R N N N N H O [Ru(bpy)2(phen-alloxazine)]2+ O H N N N Ru N H N N N H N N C H3 N N H O N O N O H N H O H R H H N O N N R N N R N O N N N N N CH3 N H N O N Ru N N N N N Ru H H N N O N N N N H N Ru N N H NH2 N N O N N N [Ru(bpy)2(phen-pterin)]2+ O Compare C-G-C triad vs. [Ru(bpy)2(phen-pterin)]-G-C triad N Ru Ru N N N O NH2 N R O H O R 2+ H N N O H N H N R CH3 [Ru(bpy)2(phen-dimethylalloxazine)]2+ Results and Discussion Fluorescence Titration of 20 M Viscosity Fluorescence Titrations Saturation Curve 5 3x10 5 2x10 5 1x10 50 100 150 [CT DNA]:[Dye] 2.0x10 5 0.0 550 600 650 700 750 Wavelength (nm) EtBr 1.15 Allox Bpy3 1.1 Diamino 1.05 DMA Pterin 1 0.95 0.00 0.05 0.10 0.15 1.72 CT DNA 1.65 1.62 1.55 1.52 Poly dAT*dAT (bpy)3 1.35 Pterin 1.3 1.25 1.2 h /ho 1/3 Diaminopteridine Alloxazine Dimethylalloxazine DPPZ 1.15 1.1 1.05 1 0.95 25 30 35 40 45 50 55 60 65 70 75 80 85 90 95 Temperature (o C) Poly dGC*dGC 1.25 CT DNA 64.5 deg C [Ru(bpy)3]2+ 64.5 deg C Ru(bpy)2pterin]2+ 64.5 deg C [Ru(bpy)2(phendiaminopteridine)]2+ 67.5 deg C [Ru(bpy)2(phenalloxazine)]2+ 75 deg C [Ru(bpy)2(phendimethylalloxazine)]2+ 78 deg C As DNA is heated, the two strands of the helix pull apart, and denatured single-stranded DNA is the result. When an intercalator is bound to the helix, it stabilizes the DNA, and Increases the melting temperature (Tm) This can be measured by recording the absorbance at 260 nm, since this is the wavelength that DNA bases absorb. With the exception of [Ru(bpy)3]2+ and [Ru(bpy)2pterin]2+, the Tm of intercalated DNA was increased compared to a shift of 64.5 to 80 oC for intercalator [Ru(bpy)2DPPZ]2+. 0.95 -0.02 Molar Ellipticity (deg cm 2 dmol-1) +Ru(bpy)3 2.0E+05 +RuDPPZ 1.0E+05 250 270 290 310 -2.0E+05 -3.0E+05 Wavelength (nm ) 10 CT DNA:Ru(bpy)2 - pterin, DMA, DAP, ALX Since surface and groove binding have little or no effect on DNA structure, intercalation can be distinguished by changes in CD signal. The bands due to base stacking at 275 nm and helicity at 245 nm are easily altered by an intercalating molecule, making circular dichroism (CD) a useful technique for determining binding mode. The top plot illustrates the changes made by a known intercalator, [Ru(bpy)2DPPZ]2+, where the positive signal centered at 275 nm in CT-DNA shifts to 270 nm and a negative band at 291 nm appears. This change is diagnostic of a B to Z transition. In contrast, the CD of non-intercalator [Ru(bpy)3]2+ added to DNA is identical to CT-DNA alone. CT DNA 2.00E+05 +Rupterin 1.50E+05 +RuDMA +RuDAP 1.00E+05 +RuALX 5.00E+04 0.00E+00 -5.00E+04230 250 270 290 -1.00E+05 -1.50E+05 -2.00E+05 Wavelength (nm) Poly dGC*dGC 1.12 0.92 -0.02 0.68 [Ru-DMA]: [base pairs DNA] [RuDiamino]: [base pairs DNA] Viscosity Titration: 0.724 mM Ru-Alloxazine in Acetonitrile & 0.14 mM DNA in buffer in Acetonitrile and DNA in buffer CT DNA (0.18 mM) 1.45 CT DNA 1.72 1.62 1.52 Poly dG*dC (0.15 mM) 1.35 1.25 CT DNA (0.09 mM) 1.15 Poly dAT*dAT 1.42 1.32 1.22 Poly dGC*dGC 1.12 1.02 0.92 1.05 0.95 -0.02 0.03 0.08 0.13 0.18 0.23 0.28 0.33 0.38 -0.02 Poly dA*dT (0.09 mM) 0.43 0.68 [RuAlloxazine]: [base pairs DNA] Preliminary experiments to test for DNA sequence specificity by the intercalating [Ru(bpy)2L]2+complexes where L= phen-alloxazine, phendimethylalloxazine and phendiaminopteridine gave the results at left. There are distinct differences among the three intercalating molecules. The phen-diamino and phendimethylalloxazine complexes bind more strongly to poly dGC-dGC than to poly dATdAT. In contrast the phenalloxazine complex binds equally well to poly dGC-dGC and poly dAT-dAT. [Ru-Alloxazine]: [base pairs DNA] Plasmid Unwinding [Ru(bpy)2(phendimethylalloxazine)]2+ 2 3 4 5 6 7 8 1 2 3 4 5 6 [Ru(bpy)2(phen-alloxazine)]2+ 7 8 1 2 3 4 5 6 7 8 [Ru(bpy)2(phendiaminopteridine)]2+ 1 2 3 4 5 6 7 8 Circular Dichroism Results CT DNA 0.0E+00 230 -1.0E+05 1.22 0.68 Viscosity Titrations: Ru-Alloxazine 1 10 CT DNA:Ru(bpy)3 & Ru(bpy)2DPPZ 3.0E+05 Poly dAT*dAT 1.32 1.02 [Ru(bpy)2(DPPZ)]2+ Circular Dichroism CT DNA 1.42 1.05 h /h o 1/3 1.4 Molar Ellipticity (deg cm 2 dmol-1) Relative Abs @ 260 nm CT DNA 1.35 1.15 Tm Values DNA:Ru 15 Viscosity Titration: 0.702 mM Ru-DMA in Acetonitrile & ~0.14 mM DNA in buffer Viscosity Titration: 0.745 mM Ru-Diamino in Acetonitrile & 0.14 mM DNA in buffer 1.45 Thermal Denaturation Results 0.20 Drug:Calf Thymus DNA 1.75 Thermal Denaturation h= viscosity t = flow time (seconds) t0 = flow time of buffer alone (seconds) h 0 = viscosity of DNA alone Dppz 1/3 0 When DNA is intercalated by a molecule, the helical structure is altered. Due to the shape distortion and lengthening of the helix, the viscosity of the DNA solution is increased. This is easily monitored by flow time of the solution. All compounds tested (see legend in plot at left), except for the known non-intercalator [Ru(bpy)3]2+ and new complex [Ru(bpy)2pterin]2+, showed significantly increased viscosity. h=(t-t0)/t0 1.2 h /ho Emission Intensity 5 1/3 Emission Intensity (cps) 4.0x10 M Viscometry Results Viscosity Titration: ~2.5 mM Drug in Acetonitrile & 0.90 mM CT DNA in buffer Initial fluorescence titrations with all four Ru(bpy)2phen-pteridine complexes showed increased emission intensity suggestive of DNA interactions. However, only three complexes produce saturation curves typical of intercalation or significant DNA interaction. These three complexes are: [Ru(bpy)2phen-alloxazine]2+, [Ru(bpy)2phen-dimethylalloxazine]2+, [Ru(bpy)2phen-diaminopteridine] 2+ A representative saturation curve is shown at left. The complex with the pterin component, [Ru(bpy)2phen-pterin] 2+, shows little increase in emission and a linear saturation plot indicating this complex does not intercalate or interact strongly with DNA. 1/3 [Dye] = 20 4x10 h /ho [Ct DNA] is Increasing 5 h /ho Ru(bpy)2phen-Alloxazine & Calf Thymus DNA 310 The CD spectra of solutions of CT-DNA with each of the four [Ru(bpy)2phen-pteridine]2+complexes indicates qualitatively the extent of intercalation. The three complexes with phen-dimethylalloxazine , phen-alloxazine, and phen-diaminopteridine show CD spectral changes indicative of intercalation while [Ru(bpy)2phen-pterin]2+shows no spectral changes from disruption of DNA structure and hence, no intercalation. Lanes 1-5: Supercoiled plasmid + increasing [Ru compound] + Topoisomerase Lane 6: Supercoiled + highest [Ru compound] Plasmid Unwinding Results Plasmid DNA is negatively supercoiled (-Lk). Upon intercalation, DNA is unwound, and Lk becomes less negative. At high concentrations of intercalator, positive supercoils (+Lk) are induced. Lane 7: Relaxed plasmid Lane 8: Supercoiled plasmid Lk: Linking number, a: Topoisomerase I relaxation, b: Ligand removal [Ru(bpy)2(phen-diaminopteridine)]2+, [Ru(bpy)2(phenalloxazine)]2+ and [Ru(bpy)2(phendimethylalloxazine)]2+ all showed plasmid unwinding behavior similar to DPPZ. Conclusions All five methods point to the same interpretation: three complexes— [Ru(bpy)2phen-alloxazine]2+, [Ru(bpy)2phen-dimethylalloxazine] 2+, and [Ru(bpy)2phen-diaminopteridine] 2+— can intercalate DNA nearly as well as ethidium bromide or [Ru(bpy)2DPPZ]2+, while [Ru(bpy)2phen-pterin]2+ shows little ability to disrupt the DNA structure and hence, no intercalation.