Improved click chemistry for EdU cell proliferation combined with GFP
fluorescence
Miguel Garcia1, Laetitia Roh1, Jessica Sordet-Dessimoz2, Gian-Filippo Mancini2, Scott Grecian3, Upinder Singh3, Kyle Gee3, Scott T.
Clarke3 1Flow Cytometry, Ecole Polytechnique Fédérale De Lausanne, 2Histology Core Facility, Ecole Polytechnique Fédérale De Lausanne, 3Cellular Imaging, Molecular
Probes/Life Molecular Probes® Labeling and Detection Technologies – Life Technologies, 29851 Willow Creek Road, Eugene, Oregon 97402 USA
RESULTS
ABSTRACT
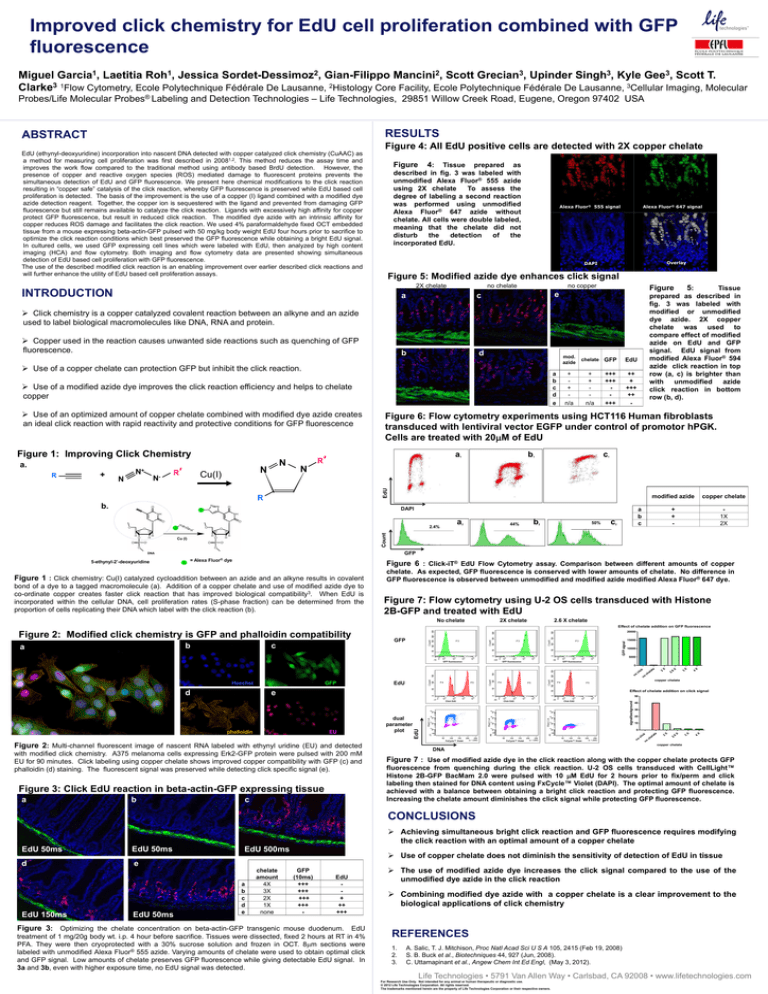
Figure 4: All EdU positive cells are detected with 2X copper chelate
EdU (ethynyl-deoxyuridine) incorporation into nascent DNA detected with copper catalyzed click chemistry (CuAAC) as
a method for measuring cell proliferation was first described in 20081,2. This method reduces the assay time and
improves the work flow compared to the traditional method using antibody based BrdU detection. However, the
presence of copper and reactive oxygen species (ROS) mediated damage to fluorescent proteins prevents the
simultaneous detection of EdU and GFP fluorescence. We present here chemical modifications to the click reaction
resulting in “copper safe” catalysis of the click reaction, whereby GFP fluorescence is preserved while EdU based cell
proliferation is detected. The basis of the improvement is the use of a copper (I) ligand combined with a modified dye
azide detection reagent. Together, the copper ion is sequestered with the ligand and prevented from damaging GFP
fluorescence but still remains available to catalyze the click reaction. Ligands with excessively high affinity for copper
protect GFP fluorescence, but result in reduced click reaction. The modified dye azide with an intrinsic affinity for
copper reduces ROS damage and facilitates the click reaction. We used 4% paraformaldehyde fixed OCT embedded
tissue from a mouse expressing beta-actin-GFP pulsed with 50 mg/kg body weight EdU four hours prior to sacrifice to
optimize the click reaction conditions which best preserved the GFP fluorescence while obtaining a bright EdU signal.
In cultured cells, we used GFP expressing cell lines which were labeled with EdU, then analyzed by high content
imaging (HCA) and flow cytometry. Both imaging and flow cytometry data are presented showing simultaneous
detection of EdU based cell proliferation with GFP fluorescence.
The use of the described modified click reaction is an enabling improvement over earlier described click reactions and
will further enhance the utility of EdU based cell proliferation assays.
Figure
4:
Tissue prepared as
described in fig. 3 was labeled with
unmodified Alexa Fluor® 555 azide
using 2X chelate
To assess the
degree of labeling a second reaction
was performed using unmodified
Alexa Fluor® 647 azide without
chelate. All cells were double labeled,
meaning that the chelate did not
disturb
the
detection
of
the
incorporated EdU.
Alexa Fluor® 555 signal
Alexa Fluor® 647 signal
Overlay
DAPI
Figure 5: Modified azide dye enhances click signal
2X chelate
INTRODUCTION
no chelate
a
c
b
d
no copper
Figure
Tissue
prepared as described in
fig. 3 was labeled with
modified or unmodified
dye azide. 2X copper
chelate was used to
compare effect of modified
azide on EdU and GFP
signal. EdU signal from
modified Alexa Fluor® 594
azide click reaction in top
row (a, c) is brighter than
with unmodified azide
click reaction in bottom
row (b, d).
e
Click chemistry is a copper catalyzed covalent reaction between an alkyne and an azide
used to label biological macromolecules like DNA, RNA and protein.
Copper used in the reaction causes unwanted side reactions such as quenching of GFP
fluorescence.
Use of a copper chelate can protection GFP but inhibit the click reaction.
c
a
b
c
d
e
Use of a modified azide dye improves the click reaction efficiency and helps to chelate
copper
Use of an optimized amount of copper chelate combined with modified dye azide creates
an ideal click reaction with rapid reactivity and protective conditions for GFP fluorescence
Figure 1: Improving Click Chemistry
R
+
N+
N-
N
R′
N
Cu(I)
chelate
GFP
EdU
+
+
n/a
+
+
n/a
+++
+++
+++
++
+
+++
++
-
Figure 6: Flow cytometry experiments using HCT116 Human fibroblasts
transduced with lentiviral vector EGFP under control of promotor hPGK.
Cells are treated with 20M of EdU
a
R′
b
1
c
1
1
N
b1
a1
EdU
a.
N
mod.
azide
R
b.
DAPI
a
2
44%
b
50%
2
c
modified azide
copper chelate
+
+
-
1X
2X
a
b
c
2
Count
2.4%
Cu (I)
GFP
DNA
5-ethynyl-2’-deoxyuridine
5:
= Alexa Fluor® dye
Figure 6 : Click-iT® EdU Flow Cytometry assay. Comparison between different amounts of copper
Figure 1 : Click chemistry: Cu(I) catalyzed cycloaddition between an azide and an alkyne results in covalent
bond of a dye to a tagged macromolecule (a). Addition of a copper chelate and use of modified azide dye to
co-ordinate copper creates faster click reaction that has improved biological compatibility3. When EdU is
incorporated within the cellular DNA, cell proliferation rates (S-phase fraction) can be determined from the
proportion of cells replicating their DNA which label with the click reaction (b).
chelate. As expected, GFP fluorescence is conserved with lower amounts of chelate. No difference in
GFP fluorescence is observed between unmodified and modified azide modified Alexa Fluor® 647 dye.
Figure 7: Flow cytometry using U-2 OS cells transduced with Histone
2B-GFP and treated with EdU
No chelate
2X chelate
2.6 X chelate
Effect of chelate addition on GFP fluorescence
b
a
c
GFP
GFP signal
Figure 2: Modified click chemistry is GFP and phalloidin compatibility
20000
15000
10000
5000
GFP fluorescence
GFP fluorescence
GFP fluorescence
Hoechst
X
X
4
2.
6
3
X
X
2
50
with modified click chemistry. A375 melanoma cells expressing Erk2-GFP protein were pulsed with 200 mM
EU for 90 minutes. Click labeling using copper chelate shows improved copper compatibility with GFP (c) and
phalloidin (d) staining. The fluorescent signal was preserved while detecting click specific signal (e).
Figure 3: Click EdU reaction in beta-actin-GFP expressing tissue
c
40
30
20
10
FxCycle™ Violet
X
4
X
3
X
X
2
2.
6
ch
no
cl
ic
k
FxCycle™ Violet
el
at
e
0
FxCycle™ Violet
Figure 2: Multi-channel fluorescent image of nascent RNA labeled with ethynyl uridine (EU) and detected
Click EdU
no
EU
dual
parameter
plot
Click EdU
EdU
phalloidin
b
el
at
e
Effect of chelate addition on click signal
e
Click EdU
a
ch
copper chelate
EdU
signal/background
d
GFP
no
no
cl
ic
k
0
copper chelate
DNA
Figure 7 : Use of modified azide dye in the click reaction along with the copper chelate protects GFP
fluorescence from quenching during the click reaction. U-2 OS cells transduced with CellLight™
Histone 2B-GFP BacMam 2.0 were pulsed with 10 M EdU for 2 hours prior to fix/perm and click
labeling then stained for DNA content using FxCycle™ Violet (DAPI). The optimal amount of chelate is
achieved with a balance between obtaining a bright click reaction and protecting GFP fluorescence.
Increasing the chelate amount diminishes the click signal while protecting GFP fluorescence.
CONCLUSIONS
Achieving simultaneous bright click reaction and GFP fluorescence requires modifying
the click reaction with an optimal amount of a copper chelate
EdU 50ms
EdU 50ms
d
e
EdU 150ms
EdU 50ms
EdU 500ms
a
b
c
d
e
chelate
amount
4X
3X
2X
1X
none
Use of copper chelate does not diminish the sensitivity of detection of EdU in tissue
GFP
(10ms)
+++
+++
+++
+++
-
EdU
+
++
+++
Figure 3: Optimizing the chelate concentration on beta-actin-GFP transgenic mouse duodenum. EdU
treatment of 1 mg/20g body wt. i.p. 4 hour before sacrifice. Tissues were dissected, fixed 2 hours at RT in 4%
PFA. They were then cryoprotected with a 30% sucrose solution and frozen in OCT. 8m sections were
labeled with unmodified Alexa Fluor® 555 azide. Varying amounts of chelate were used to obtain optimal click
and GFP signal. Low amounts of chelate preserves GFP fluorescence while giving detectable EdU signal. In
3a and 3b, even with higher exposure time, no EdU signal was detected.
The use of modified azide dye increases the click signal compared to the use of the
unmodified dye azide in the click reaction
Combining modified dye azide with a copper chelate is a clear improvement to the
biological applications of click chemistry
REFERENCES
1.
2.
3.
A. Salic, T. J. Mitchison, Proc Natl Acad Sci U S A 105, 2415 (Feb 19, 2008)
S. B. Buck et al., Biotechniques 44, 927 (Jun, 2008).
C. Uttamapinant et al., Angew Chem Int Ed Engl, (May 3, 2012).
Life Technologies • 5791 Van Allen Way • Carlsbad, CA 92008 • www.lifetechnologies.com
For Research Use Only. Not intended for any animal or human therapeutic or diagnostic use.
© 2012 Life Technologies Corporation. All rights reserved.
The trademarks mentioned herein are the property of Life Technologies Corporation or their respective owners.