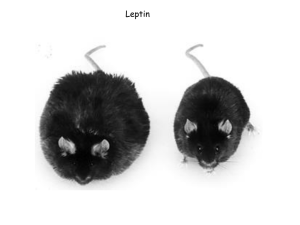
Improving Physicochemical Properties of Biopharmaceutical Drug
advertisement

Improving Physicochemical Properties of Biopharmaceutical Drug Candidates David Litzinger, PhD Director, Pharmaceutical Sciences Amylin Pharmaceuticals, Inc. PEGS Conference May 20, 2010 Boston, MA Analogs in Drug Development Comparisons Across Platforms Drug Platform (typical MW) Analog Evaluation in Drug Development Immunogenicity Concern Small Molecules High Negligible Medium Slight Low Significant <500 Da Peptides 1-6 kDa Proteins 15-150 kDa Peptide Analogs as Drug Substances Examples Related to Aggregation Drug Substance Endogenous Counterpart Mutations Result pramlintide amylin • Three proline substitutions • Prevents insoluble fibrous aggregate formation • Based on rat amylin (has the three corresponding prolines, is not amyloidogenic) Insulin glargine insulin • Two Arg added to B chain (shifts pI from 5.4 to 6.7) • Gly to Asn at A21 Insulin lispro Insulin aspart insulin insulin • Formulated as a solution at acidic pH • Following injection, comes out of solution at physiological pH to form crystals that slowly dissolve • Lys and Pro at the C-terminal end of the B-chain reversed • Blocks the formation of insulin dimers and hexamers • Pro to Asp at B28 • Increased charge repulsion prevents the formation of hexamers • Rapid acting insulin • Rapid acting insulin Insulin glulisine insulin • Asn to Lys at B3 • Lys to Glu at B29 • Rapid acting insulin Peptide and Protein Optimization Example Options for Improving Physical Stability Approaches to Improving Physical Stability* Mutational Chemical Modification – Mutations based on superior properties in alternate species √ – Decrease hydrophobicity √ √ – Increase hydrophilicity √ √ – Increase net charge √ √ – Changing the pI √ √ – Polymer conjugation * More that one approach can be combined √ Glucose-Dependent Insulinotropic Polypeptide Example of Analog Evaluation in Drug Development > Glucose-dependent Insulinotropic Polypeptide (GIP) – 42-amino acid hormone synthesized and secreted from intestinal K-cells – Integral role in regulating insulin secretion and response – Amylin Pharmaceuticals currently investigating GIP as a possible mono- or combination therapy for Type 2 Diabetes Mellitus > Development Challenges – Native GIP rapidly inactivated by dipeptidyl peptidase-IV (DPP-IV) and has a very short half-life – Development of GIP analogs challenging due to poor solubility > Second Generation Effort (G2) – G1 effort addressed DPP-IV metabolism, optimized activity – G2 GIP analogs identified and evaluated for improved solubility • In Silico modeling used for primary sequences analysis • pH-solubility profile, physical and chemical stability were screened • CD used to monitor secondary structure GIP Drug Development History and Efforts to Identify Alternative GIP Analogs Generation Peptide ID# Metabolism Biological Activity Physical Stability P Human GIP (1-42) X X G1 G1-A G1 G1-B G2 G2-C G2 G2-D x x Note: Biological Activity- Receptor binding, mouse OGTT, mouse GL, DOA by rat IVGTT, plasma stability, HbA1c in ob/ob mice Physical Stability- Aggregation, precipitation, solubility Native GIP (1-42) G1 Analogs 2nd Round of Screening G2 Analogs GIP Analog Screening Primary Sequence Ranking by In Silico Tools Generation Peptide ID# P Primary Sequence Human GIP YAEGTFISDYSIAMDKIHQQDFVNWLLAQKGKKNDWKHNITQ-OH MW 4983.6 G1 G1-A YaEGTFISDYSIAMDKIHQQDFVNWLLAQKPSSGAPPPS-NH2 4309.8 G1 G1-B YaEGTFISDYSIAMDKIHQQDFVNWLLAQKPSSGAPPNS-NH2 4326.8 G2 G2-C YaEGTFISDYSIALEKIRQQEFVNWLLAQKPSSGAPKPS-NH2 4369.9 G2 G2-D YaEGTFISDYSIALEKIRQQEFVNWLLAQKPSSGAPPKSK-NH2 4498.1 Underlined residues denote substitutions; Red - potentially labile residues; Blue – C-terminal end modification > Sequences ranked according to In Silico modeling and assessment tools – Tango2 – Protein aggregation prediction model based on TANGO algorithm of physico-chemical principles of b-sheet formation – In Silico Tool – Primary sequence assessment and pharmaceutical properties predictor created in-house • GRAVY– Grand average of hydropathicity: GRAVY value, hydrophobicity ( solubility) • Peptide Charge Calculator – Computes theoretical net charge on peptide from composition of ionizable residues > Compounds synthesized and evaluated GIP Analog Screening In Silico Pharmaceutical Property Assessments Solubility ID # Human GIP Hydrophilicity Aggregation (Gravy Score) (Tango 2 Score) Calculated pH 4 pH 7 Net Charge Net Charge Overall pI Solubility Solubility pH 4 pH 7 Stability Good + 5.68 + 0.39 + 2.86 - 0.85 Good D(3), M(1), N(1), Q(3), W (1) + 2.86 - 0.85 Good D(3), M(1), N(2), Q(3), W (1) + 3.86 + 0.91 Good D(1), N(1), Q(3), W (1) + 4.86 + 1.91 Good D(1), N(1), Q(3), W (1) -0.80 -7.00 7.5 G1-A -0.37 -7.10 5.8 G1-B -0.42 -6.78 5.8 G2-C -0.41 -13.89 8.6 Good G2-D -0.50 -14.57 9.2 Good (1-42) Chemical Stability Fair Average Fair Fair Average Average Fair Fair Average Average Fair Average Fair Average Fair Average Potential Labile Residues D(4), M(1), N(3), Q(4), W (2) > G2 analogs showed improved properties over G1 analogs: • Higher pI • Good solubility at acidic pH • Fair/Average solubility at neutral pH • Highly charged at pH 4 compared to pH 7 > Labile Residues: • D – potential aspartic acid isomerization at pH 4 • M, W – potential site for oxidation • N, Q – potential deamidation Measured Solubility Results G2 Analogs Have Improved Solubility ID # Solubility at Formulated pH Hydrophilicity (Gravy Score) Aggregation (Tango 2 Score) Measured pI Calculated pI Human GIP (1-42) nd -0.80 -7.00 6.7 7.5 G1-A < 1 mg/ml -0.37 -7.10 5.8 5.8 G1-B ~ 1 mg/ml -0.42 -6.78 4.7 5.8 G2-C > 5 mg/ml -0.41 -13.89 8.4 8.6 G2-D > 5 mg/ml -0.50 -14.57 9.0 9.2 Note: nd – not determined > G2 analogs show improved solubility profile compared to the G1 analogs Formulation Screening G2 Analogs Have Improved Physical Stability ID # Temperature at 25°C pH G1-A Buffer 5.0 30 mM Acetate 6.0 30 mM Phosphate 6.0 30 mM Histidine 6.5 30 mM Phosphate 7.0 30 mM Phosphate 5.0 30 mM Acetate 6.0 30 mM Phosphate 6.0 30 mM Histidine 6.5 30 mM Phosphate 7.0 30 mM Phosphate 6.0 10 mM Phosphate 6.5 10 mM Phosphate 6.5 10 mM Histidine 7.0 10 mM Phosphate 7.0 30 mM Phosphate 7.0 10 mM Histidine 7.5 10 mM Phosphate 5.0 10 mM Acetate 5.5 10 mM Acetate 6.0 10 mM Histidine 6.5 10 mM Histidine 7.0 10 mM Histidine 7.5 10 mM Histidine Time Point (Weeks) 0 1 2 Visual Analysis Clear, Colorless Slight Precipitation, Aggregation G1-B G2-C G2-D Moderate to Severe Precipitation, Aggregation > G2 analogs proved to have the most physically stable profile. 1 mg/mL concentration; No agitation Secondary Structure Analysis Evaluation of GIP Analogs Far FarUV UVCD CD Mean Residue Ellipticity (MRE) (mdeg*(cm2/dmol) 25000 20000 Structure l (nm) 15000 α-helix 208, 220 10000 β-sheet 215 Random Coil 195 5000 G1-A pH 6 Phosphate G1-B pH 6 Phosphate G2-C pH 4 Acetate 0 G2-C pH 7 Phosphate -5000 -10000 G2-D pH 4 Acetate -15000 -20000 190 G2-D pH 7 Phosphate 200 210 220 230 Wavelength (nm) 240 250 > G2 analogs show greater α-helical content – – Correlates with less aggregation Similar 2° structure at both pH 4 & 7 260 GIP Analog Optimization Conclusions > G1 analogs demonstrated improved biological efficacy and longer duration of action compared to native GIP, but had poor physical stability > G2 analogs showed both improved efficacy and physical stability – Experimental results correlated well with their higher net charge and more negative GRAVY scores predicted in silico. – At 1 mg/mL concentrations were physically and chemically stable under the tested conditions with little to no visible aggregation. – Secondary structure is predominantly α-helical in liquid state (pH 4.0 and pH 7.0) Metreleptin Compound Properties and Obesity Treatment Approaches • 16.2 kd 147 amino acids, (native leptin 146 AA) • Isoelectric point 6.1 • Single disulfide bond • No free cysteines • Limited solubility at neutral pH, 2-3 mg/mL, higher at lower pH • Four helix bundle tertiary structure > Amgen pursued leptin monotherapy as a treatment for obesity – High dose, up to 0.3 mg/kg (~30 mg per injection) – Heymsfield et al. (1999) JAMA > Amylin is evaluating leptin in combination with pramlintide for treatment of obesity – Lower dose – Roth et al. (2008) PNAS Metreleptin Charge Profile Net Charge of Metreleptin vs pH > Calculated pI= 6.1 > Suggests high solubility at low pH, and low solubility at neutral pH Charge calculator/pI finder by Gale Rhodes http://spdbv.vital-it.ch/TheMolecularLevel/Goodies/Goodies.html Metreleptin Solubility Profile ○ leptin solubility ▲ reversibility of precipitation > Solubility of human leptin – At low pH is high > 70 mg/mL at pH 4 – At neutral pH is low 2-3 mg/mL > Precipitation at neutral pH is essentially irreversible > Murine leptin is more soluble than human leptin at neutral pH – 43 mg/mL for murine leptin – 31 mg/mL for W100Q/W138Q analog Ricci, M.S. et al. (2006) in Misbehaving Proteins. New York: Springer. Human and Murine Leptin Amino Acid Sequence Comparison > Comparison of human and murine leptin sequences 0 HUMAN: MVPIQKVQDD MURINE: MVPIQKVQDD 40 HUMAN: DFIPGLHPIL MURINE: DFIPGLHPIL 80 HUMAN: LENLRDLLHV MURINE: LENLRDLLHL 120 HUMAN: STEVVALSRL MURINE: STEVVALSRL 10 20 30 TKTLIKTIVT TKTLIKTIVT RINDISHTQS RINDISHTQS VSSKQKVTGL VSAKQRVTGL 50 60 70 TLSKMDQTLA SLSKMDQTLA VYQQILTSMP VYQQVLTSLP SRNVIQISND SQNVLQIAND 90 100 110 LAFSKSCHLP LAFSKSCSLP WASGLETLDS QTSGLQKPES LGGVLEASGY LDGVLEASLY 130 140 QGSLQDMLWQ QGSLQD I LQQ LDLSPGC LDVSPEC Residues that differ between the human and murine sequences are in red. Note that the first methionine residue associated with E. coli production is not counted. – Differ at 22 sites – Sequence differences of particular significance in solubility/aggregation properties Metreleptin Surface Modeling Electrostatic Surface Red = Basic (+) Blue = Acidic(-) Hydrophobicity Surface Brown = Lipophilic Blue = Hydrophilic, charged Trp 138 > Surface modeling shows region around Trp 138 has potential role in aggregation – Low electrostatic potential – High lipophilicity Benchware3DExplorer (Tripos) Human Leptin Evidence for Leptin Conformational Transition with pH Change ● human ○ murine > Increased ANS fluorescence at pH 4 to 5 – Not observed for murine leptin > Suggests a folding intermediate with increased hydrophobicity populated at pH 4-5 > May result in the formation of soluble multimeric species under acidic conditions Ricci, M.S. et al. (2006) in Misbehaving Proteins. New York: Springer. Human Leptin Low pH Aggregation and Relation to Neutral pH Precipitation ▲ human, % aggregates, pH 4 ● human, % precipitation, pH 7 ∆ murine, % aggregates, pH 4 ○ murine, % precipitation, pH 7 Initial concentration at low pH varied Precipitation induced by diluting into neutral pH buffer Inset: human leptin multimers formed at 50 mg/mL, pH 4: • diluted to 5 mg/mL, pH 4 • diluted again into pH 7 Human leptin Forms multimers at low pH Precipitation correlates with multimer formation Multimers formed at acidic pH dissociate upon dilution in acid pH Precipitation at pH 7 decreases with multimer dissociation Murine leptin Did NOT form multimers and did not precipitate Ricci, M.S. et al. (2006) in Misbehaving Proteins. New York: Springer. Human Leptin Proposed Aggregation Mechanism Increased hydrophobicity at acidic pH not observed** N Multimers not observed* Murine Leptin I U Iassoc Precipitation not observed* precipitation * Under conditions in which human leptin precipitated, and formed multimers. N: native state I: folding intermediate ** As observed for human leptin in ANS studies. U: unfolded conformer Iassoc: folding intermediate self associated into a soluble multimer Ricci, M.S. et al. (2006) in Misbehaving Proteins. New York: Springer. Chemical Modification Example Succinylation O O Protein-NH2 + O Protein-N-C-CH2-CH2-C-OH2 O > Reaction at pH 7.0 – 5-fold molar excess of succinic anhydride – 2-16 hours at 4oC > Purification by ion exchange chromatography – 45-47% final yield > Site-specific conjugation to N-terminus – Endoproteinase Lys-C – Peptides resolved by RP-HPLC • M1-K6: N-terminal peptide From Gegg et al. US Patent 6,420,340 O • Succ-(M1-K6): Succinylated N-terminal peptide Two Related Examples DTPA and EDTA O O O N-R-N O H2N-Protein R = -CH2-CH2-N-CH2CH2H+ (2) O O O HO OH OH HO O O N-Protein H CH2 H2O (1) or H2N-Protein (2) (1) O COOH O O N-R-N Diethylenetriaminepentaacetic acid (DTPA) Monomer conjugate From Gegg et al. US Patent 6,420,340 O OH N-R-N N-Protein H O N-Protein H Dimer conjugate Ethylenediaminetetraacetic acid (EDTA) R = -CH2-CH2- Succinylation and Related Modifications Impact on pI and Solubility of Metreleptin Sample Maximum Solubility in PBS* (mg/mL) Change in pI Unmodified leptin Succinyl-leptin 3.2 8.4 N/A ** -0.7 ** DTPA-leptin monomer EDTA-leptin monomer 31.6 59.9 Not reported Not reported * pH = 7.0 ** Leptin pI = 6.1; succinyl-leptin estimated to be 5.4 > Conjugations with acidic moieties to the N-terminus lower pI and increase solubility at neutral pH From Gegg et al. US Patent 6,420,340 Succinylation Reduces Injection Site Precipitation of Metreleptin > Three mice dosed per sample > Tissues sections from the injection sites examined histologically Sample Concentration (mg/mL) Acetate buffer, pH 4.0 Unmodified leptin (in acetate buffer, pH 4.0) PBS buffer, pH 7.5 Succinyl-leptin (in PBS, pH 7.5) 0 0 0 50 50 50 0 0 0 50 50 50 Injection volume (mL) 20 20 20 20 20 20 20 20 20 20 20 20 Score system: 0 Normal, 0.5-1 minimal change, 1.5-2 mild change, 2.5-3 moderate change, 3.5-4 marked change, 4.5-5 massive change From Gegg et al. US Patent 6,420,340 Precipitation 0 0 0 4 4 1.5 0 0 0 0 0.5 0 Succinylated and Related Metreleptin Conjugates Retain In Vivo Activity > Similar activity in vivo for conjugates relative to unmodified leptin – Normal mice dosed s.c. daily, 10 mg/kg – Results shown as % weight-loss relative to buffer control From Gegg et al. US Patent 6,420,340 Polymer Conjugation Example PEGylation > What is PEGylation? – Covalent attachment of poly(ethylene glycol) (PEG) – Example PEGylation reagent: Methoxy cap CH3O-(CH2-CH2-O)n-CH2-CH2-X Reactive group > Why PEGylation? – Slow clearance/maintain circulating concentrations/reduce dose frequency – Increase solubility – Reduce aggregation – Reduce proteolysis – Reduce immunogenicity – In several approved products Site-Directed PEGylation N-Terminal Site-Specific Example NeH3+ -OOC NeH3+ O H-C-PEG Protein -OOC NH2 Protein NaCNBH3 NeH3+ NeH3+ – Low pH selectively protonates lysine e-amino groups – N-terminal amino group remains unprotonated and reactive – Reductive alkylation specific to the N-terminus Example: Neulasta® (20kDa PEG-rhGCSF) > Why site-directed PEGylation? – Optimally preserve biological activity – Homogenous product/consistent lot-to-lot activity NH-CH2-PEG Effect of PEGylation on Solubility PEG-GCSF Has Improved Solubility > Under conditions in which GCSF rapidly precipitated, 20kDa PEG-GCSF remained completely soluble Samples formulated at 5 mg/mL in phosphate buffer, pH 6.9 and incubated at 37oC > PEG-GCSF remained clear and showed no turbidity, unlike GCSF > Free PEG was unable to prevent GCSF precipitation From Rajan, R.S. et al. (2006) Protein Science PEG-GCSF Forms Soluble Aggregates Analysis by Size-Exclusion Chromatography > Significant loss of GCSF monomer due to conversion into insoluble forms > 20K PEG-GCSF accumulated into soluble, higher order multimeric forms eluting in the void volume * Aliquots analyzed after 72 h of incubation at neutral pH and 37oC From Rajan, R.S. et al. (2006) Protein Science PEGylation and Aggregation Mechanism Findings > PEGylation does not alter the linkages or heterogeneity of the aggregates – Resolubilized GCSF and PEG-GCSF soluble aggregates comparison • Both included a mixture of monomer, dimers, trimers, and higher order multimers • Multimers in both cases were covalent, disulfide-linked • Similar extent of covalent formation > PEGylation does not alter the helix-to-sheet transition that accompanies aggregation – GCSF and PEG-GCSF showed similar starting FTIR spectral profiles as well as temperature-induced conversion to b-sheet – The GCSF precipitate and the PEG-GCSF soluble aggregate showed similar extent of b-sheet content by FTIR analysis > PEGylation confers improved solvation by water molecules – In phase partition studies, GCSF aggregates partitioned to octanol while PEG-GCSF aggregates remained in the aqueous phase From Rajan, R.S. et al. (2006) Protein Science Aggregation and Drug Development Improving the Drug Compound > Identify potential issues early – Dose level, dose concentration – Solubility at physiological pH – Manufacturing, shipping and handling > Consider strategy to reduce aggregation – Remove aggregates during manufacture – Formulate to prevent aggregate formation – Modify the compound to reduce/remove aggregation potential > Generally, testing compounds early is preferred – Logistical benefit, test compounds while in vitro and in vivo screens are in process (rather than restarting assays) – Opportunity to solve before Candidate Nomination Early Pharmaceutical Development Opportunities to identify and solve aggregation issues during SAR development Stage 1 Stage 3 Stage 2 • Analytical method development • Analytical method optimization • Early screening • Late screening • IND enabling • Phase I enabling • Developability risk assessment Team formation Compound screening Pre-project activities • In silico modeling Candidate nomination IND Phase I activities • Monitor • Address questions/issues Acknowledgments and References Acknowledgments Pharmaceutical Sciences Steven Ren Derrick Katayama Ellen Padrique Johnny Gonzales Jenny Jin Biology Diane Hargrove Eric Kendall Augustine Cho Krystyna Tatarkiewicz Slave Gedulin Biology, cont’d Pam Smith Christine Villescaz Tina Whisenant Lynn Jodka Kim DeConzo Julie Hoyt Jenne Pierce Amy Carroll Aung Lwin Bioanalytical Chemistry Swati Gupta Kristine De Dios Liying Jiang References M.S. Ricci et al. (2006) Mutational Approach to Improve Physical Stability of Protein Therapeutics Susceptible to Aggregation. In Misbehaving Proteins (Murphy RM and Tsai AM, ed) pp331-350. New York: Springer. Gegg, C. and Kinstler, O. (2002) Chemical modification of proteins to improve biocompatibility and bioactivity. US Patent 6,420,340 Rajan, R.S. et al. (2006) Modulation of protein aggregation by polyethylene glycol Conjugation: GCSF as a case study. Protein Science 15: 1063-1075. Informatics Eugene Coats Robert Feinstein Paul Nelson Research Chemistry Odile Levy Ramina Nazarbaghi Lawrence D’Souza John Ahn