Topic 7 Phylogeny
advertisement

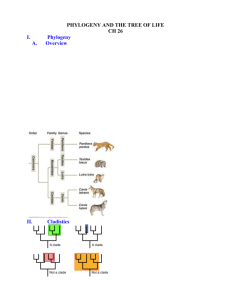
Introduction to Phylogeny Required Reading: chapter 4 (Ignore Box 4.1) Objectives • The basics of phylogenetic trees • How phylogenetic trees are constructed. • How phylogenies can address questions about evolution. Phylogeny Phylogenetics: the study of ancestor descendent relationships. The objective of phylogeneticists is to construct phylogenies Phylogeny: A hypothesis of ancestor descendent relationships. Phylogenetic tree: a graphical summary of a phylogeny Phylogeny All life forms are related by common ancestry and descent. The construction of phylogenies provides explanations of the diversity seen in the natural world. Phylogenies can be based on morphological data, physiological data, molecular data or all three. Today, phylogenies are usually constructed using DNA sequence data Review: Introduction to Phylogenetic Trees Phylogenetic Characters We use characters to construct phylogenies. A character is any attribute of an organism that can provide us with insights into history (shared ancestry). In molecular phylogenies, characters are typically nucleotide positions in a gene sequence, and each position can possess four CHARACTER STATES: A,C, G, or T Cladisitc Character State Definitions • Plesiomorphy: refers to the ancestral character state • Apomorphy: a character state different than the ancestral state, or DERIVED STATE • Synapomorphy: a derived character state (apomorphy) that is SHARED by two or more taxa due to inheritance from a common ancestor: these character states are phylogenetically informative using the parsimony or cladistic criterion • Autapomorphy: a uniquely derived character state Character States we will return to this After Page and Holmes 1998 More Synapomorphies shared shared Figure 4.2, pg.113 Definitions Monophyletic: a group that includes ALL of the descendents of a common ancestor. Monophyletic groups are also known as CLADES Non Monophyletic: Any case that does not satisfy the above, such as: Paraphyletic: A group that includes some, but not all of the descendents of a common ancestor Polyphyletic: assemblages of taxa that have been erroneously grouped on the basis of homoplasious characters (eg “vultures”) Monophyletic Groups fig 4.1 p112 •All groups circled in red are monophyletic Examples of Synapomorphies Synapomorphies identify monophyletic groups Figure 4.3, pg. 114 Monophyly and Non-Monophyly After Page and Holmes1998 Reptiles: A Paraphyletic Group Naming based on past data Naming based on current data Reptiles Sauropsida Paraphyletic – a grouping that contains some, but not all descendants of a common ancestor Homology and Homoplasy A character state that is shared between two DNA sequences or taxa may be so because they inherited it from a common ancestor, or it is HOMOLOGOUS (a homology/ synapomorphy) Alternatively, the shared character might occur because they were evolved independently, in which case they are called a HOMOPLASY Why can Homoplasy Occur? After Page and Holmes1998 Homoplasy and Polyphyly Homoplasy results in erroneous, polyphyletic groupings such as “vultures” A ‘Vulture’ “Vultures” are a polyphyletic group. New world and old world vultures provide an example of homoplasy resulting from convergent evolution. More Examples of Convergence Analogy (non homology): The fins of a whale and the fins of a shark are another example of homoplasy due to convergence, the independent acquisition of a character in different lineages Three Spine Stickle Back: Parallel Evolution • 3 spine stickle back species pairs have evolved independently in coastal lakes of British Columbia • Positive assortative mating and disruptive selection have been important in the divergence of these pairs Reversals & Phylogeny Figure 4.5, pg. 116 Constructing Phylogenetic Trees • We use homologous characters (synapomorphies) to construct phylogenetic trees and to identify groups that are monophyletic; synapomorphies are phylogenetically informative. • We want to avoid using homoplasious characters to construct phylogenies Homology and Homoplasy Revisited Parsimony (also known as cladistics) The Principle of Parsimony: simple explanations are preferred over more complicated ones. In terms of phylogenetic trees, less evolutionary steps are better than more steps to explain relationships. The tree with the least number of steps is the most parsimonious. The parsimony method minimizes the total number of evolutionary changes required to explain relationships Making Inferences With Parsimony: Evolution of the Camera Eye Constructing Trees with Parsimony Outgroup: When constructing a phylogeny for a group of organisms, we need to employ an outgroup, which is not part of the group of interest (the ingroup), but also not too distantly related to it. The outgroup is used to polarize the character states, or infer change. The character state possessed by the outgroup is defined a priori as ancestral (pleisiomorphic) Whale Evolution Ambulocetus The Artiodactyla The artiodactyla are a group of hoofed mammals that possess an even number of toes, and includes camels, pigs, peccaries, deer, the hippopotamus, cattle and giraffes. The perissodactyla are hoofed mammals that possess an odd number of toes (e.g. horses, rhinos, tapirs). Are whales really a member of the artiodactyla? Selecting Phylogenetic Trees with Parsimony Whales early Figure 4.8, pg. 121 Whales late Parsimony using morphology Figure 14.5, pg. 558 Outgroup is a Perissodactyl Parsimony using molecular characters Figure 14.6, pg. 559 • Site 142 is plesiomorphic (uninformative) • Site 192 is a autapomorphic (uninformative) • Sites 162, 166 & 177 are synapomorphies (informative) What do the informative sites tell us about whale phylogeny? • Site 162 & 166 conflict with site 177 • Hence there is homoplasy in the data set. • What is the most parsimonious tree looking at all characters? – Whales early – 47 nt changes – Whales late – 41 nt changes •Whales late has less evolutionary steps to explain relationships: the most parsimonious explanation Assessing Confidence in Phylogeny • Bootstrap Method – Computational technique for estimating the confidence level of a phylogenetic hypothesis. • Randomly generates new data sets from the original set (1000 replicates is most common) • Computes the number of times that a particular grouping (or branch) appeared in the tree. Phylogeny and Taxonomy • Taxonomic groups can be: – Monophyletic – contain all descendants of a common ancestor – Paraphyletic – contain some but not all descendants of a common ancestor, or polyphyletic (erroneous homoplasious groupings) – The goal of cladistic taxonomy is to only recognize monophyletic groups as valid taxa, but traditional taxonomy has not always done this • Cladistics- the use of parsimony to construct evolutionary relationships • cladistic taxonomy= evolutionary taxonomy Basics of Taxonomy Three domains of life: Archaea, Bacteria, Eukarya. The Eukarya are hierarchically divided as follows: Super group Kingdom Phylum Class Order Family Genus Species Unikonta Animalia Chordata Mammalia Primata Hominidae Homo sapiens Example: The Amniota All aminotic eggs possess several membranes (the amnion, chorion and allantois) that protect the developing embryo. The amniotic egg was an important evolutionary innovation and adaptation for life on land, and protects the developing embryo from desiccation Paraphyletic Groups: many taxonomic groups that were recognized by traditional taxonomy are paraphyletic (eg fish) Prokaryotes, Fish and Dicots (in addition to ‘reptiles’) are all examples of paraphyletic groups Based on past data Based on current data Artiodactyla Cetartiodactyla The Artiodactyla are another example of a paraphyletic grouping Using Phylogenies: Chameleons Biogeography is the branch of science that seeks explanations for why organisms are found in some regions, but not others. This very often involves the use of phylogenies to test hypotheses concerning the geographic origins of different species, or groups of species such as the Chameleons (we will consider biogeography in much more detail later in the course) Using Phylogenies: Chameleons Using Phylogenies: Coevolution Coevolution: The process where evolutionary changes in the traits of one species drives evolutionary changes in the traits of another species. Coevolution can involve predators and prey, hosts and parasites, and mutualisms, such as aphids and their endosymbiotic bacteria (above). Coevolution can result in co-speciation. Phylogeny & Coevolution Figure 4.17, pg 136 Other Phylogenetic Methods We have discussed the method of Parsimony, or Cladistics in phylogenetic reconstruction. However, other more powerful methods are available for use with DNA sequence data. These are collectively referred to as frequency probability methods, and include Maximum Likelihood, and Bayesian methods of phylogenetic inference. These are computationally intensive, and have only been in frequent use for the past 12 years or so, when computers became powerful enough to accommodate them These methods are covered in Biol 366 and Biol 480 Phylogeny Summary • We must use characters that are homologous (synapomorphies) and avoid homoplasies in phylogeny construction • Parsimony seeks the simplest explanation that requires the least amount of change (fewest steps). • Phylogenetic reconstruction is a powerful tool that can be used to answer many evolutionary questions