Lecture5
advertisement

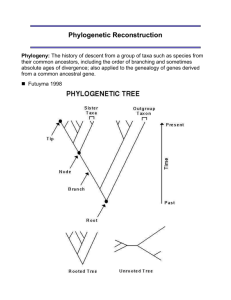
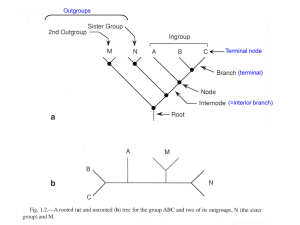
Molecular Phylogenetics Phylogenetic trees are about visualizing evolutionary relationships “Nothing in Biology Makes Sense Except in the Light of Evolution” Theodosius Dobzhansky (1900-1975) Phylogeny Hypothesis of evolutionary relationships Phylogenetic tree = graphical summary of evolutionary history We have been using trees throughout the semester Now we will examine how to construct them Phylogeny is only an estimate Phylogenetics Under Darwin’s hypothesis of common descent Species in the same genus stem from a recent ancestor Hierarchical classification reflects not a mystical ordering of the universe, but rather a real historical process Phylogenies Species tree (how are my species related?) contains only one representative from each species when did speciation take place? all nodes indicate speciation events Gene tree (how are my genes related?) normally contains a number of genes from a single species nodes relate either to speciation or gene duplication events Phylogenetic Trees Diagram consisting of branches and nodes A B C D E terminal node interior node split (bipartition) also written AB|CDE or portrayed **--- branch (edge) root of tree Unrooted vs. rooted trees Rooting a Phylogeny Several methods used to identify polarity Most commonly used is the outgroup method The character state of the target taxa is compared with that of a relative that diverged earlier Outgroup represents the ancestral state Identify outgroup from other phylogenetic studies or fossil data Good to use several outgroups at once Rooting Using an Outgroup 1. The outgroup should be a sequence (or set of sequences or taxon) known to be less closely related to the rest of the sequences (taxa) than they are to each other 2. It should ideally be as closely related as possible to the rest of the sequences (taxa) while still satisfying condition 1 The root must be somewhere between the outgroup and the rest (either on the node or in a branch) The POINT of rooting (using an outgroup) is to include the ancestor of the group of interest in the phylogeny! Terms Clade: A set of species (or sequences) which includes all of the species (or sequences) derived from a single common ancestor Monophyly Polyphyly Paraphyly Cladograms VS. Phylograms Cladogram Only shows you the relationships between taxa Branch lengths provide no data! Phylogram Shows you relationships AND the amount of change (evolution) inferred along each branch Therefore, branch lengths are very important! Cladogram Phylogram [sometimes Phenogram] (branch lengths mean something) Cladograms VS. Phylograms Species A Species A Species B Species B Species C Species C Species D Species E Species D Species E Species F Species F 5 changes Phylogenetics Terms Monophyletic Group Paraphyletic Group All members are believed to stem from a single common ancestor, and the group includes this common ancestor Group that is monophyletic except that some descendents of the common ancestor have been removed Polyphyletic Group consisting of unrelated lineages, each more closely related to other lineages not placed in the taxon Cladistic Methods Techniques that identify monophyletic groups based on synapomorphies Synapomorphies define evolutionary branching points Autapomorphies and ancestral characters do not Must be able to identify homology of traits and direction of change through time (Polarization) Homology The features of organisms almost always evolve from pre-existing features of their ancestors Unlikely that features arise de novo from nothing… Homology Homologous features are derived from a common ancestor Organs of 2 organisms are homologous if they have been inherited (& perhaps modified) from a single organ of a common ancestor A character may be homologous among species but a character state may not 5 toed state is homologous in humans and lizards but the 3 toed state is not homologous in Guinea pigs and Sloths The wings of birds and those of bats are not homologous, although their forelimbs in general are homologous structures (convergent evolution) Maximum Parsimony (Cladistic) Occam’s Razor Entia non sunt multiplicanda praeter necessitatem. William of Occam (1300-1349) The best tree is the one which requires the least number of substitutions Parsimony and Phylogeny Most closely related taxa should have the most traits in common Assume that traits are independent, heritable, and variable in target taxa Traits may be DNA sequence, presence or absence of skeletal elements or floral parts, mode of embryonic development, etc. Traits scored in different taxa must be homologous Parsimony and Phylogeny Shared derived characters (ONLY) are used to deduce the branching patterns of the tree Synapomorphy Synapomorphies are used to attach two branches at a NODE on the tree Molecular Synapomorphies Molecular Synapomorphies Parsimony and Phylogeny Traits may revert to ancestral form because of mutation or selection This may destroy phylogenetic signal and lead to reconstruction of misleading relationships Reversal Convergence and Reversal and collectively known as Homoplasy Molecular Homoplasy via Reversal Parsimony and Phylogeny Homoplasy Creates noise in the data Some characters give conflicting information about relationships Systematists try to minimize homoplasy in a data set Choose characters that evolve slowly relative to age of taxa Parsimony and Phylogeny Parsimony minimizes total amount of evolutionary change in a tree Synapomorphies are usually more common than convergence and reversal Most parsimonious trees minimize homoplasy to give best estimate of phylogeny Fitch (equal-weighted) parsimony Data for site 1 shown on tree topology for all 16 possible combinations of states at the 2 interior nodes. Character length is 2 for this site. Tree length (or tree score) Total steps = 2 + 1 + 2 + 2 + Character length from site 1 C A B A B B D C D D C 225 (best) + 1 = 237 Character length from site 2 A 237 ... 241 (worst) This value is used to compare this tree topology to other tree topologies (smaller is better) Phylogenetic Characters Which characters should be used to reconstruct the correct phylogeny? Morphological characters ie, Skeleton For fossils only morphological characters can be used Morphological characters difficult to use because taxonomic expert needed Molecular characters Allozymes, RFLPs, DNA sequences MUST CHOOSE MOLECULAR MARKER THAT IS APPROPRIATE Best molecular marker is one which has plenty of variation (=phylogenetic signal) yet not too much homoplasy (not too variable!). Phylogenetic Characters Which characters should be used to reconstruct the phylogeny? Molecular data has the advantage that they can be rapidly collected and scored However, homoplasy difficult to indentify Only four bases: G, A, T, C Multiple types of data (including multiple gene sequences) often the best What sequences should I use for organism phylogenies? Slowly evolving / Fast evolving rRNA mitochondrion Nuclear chloroplast Other Phylogenetic methods Parsimony is not the only method for estimating phylogenetic relationships!… Some pitfalls of Parsimony… It can take quite a long time to compute a Parsimony estimate of a phylogeny… Also, parsimony may be very error prone when: rates of evolution are variable very divergent species (or OTUs) are compares because it does not deal well with accounting for homoplasy… Other Phylogenetic methods Other reconstruction methods Distance (Phenetic) methods e.g.: Neighbor joining and UPGMA Based on clustering technique Based on overall similarity Not a cladistic method Uses differences (distances) among character states to group taxa Using Distance Methods to Reconstruct Phylogenetic Relationships Species with the LEAST genetic distance (or other distance) between them are assumed to be CLOSE relatives However, there are MANY cases where this may NOT be true! Distance-Based Methods (UPGMA, Neighbor Joining, etc..) Distance methods are typically very very fast and easy to use to estimate a phylogenetic tree However, they are not cladistic because they do not look for synapomorphies, but rather overall similarity… This means this method is also susceptible to lots of error when a dataset has lots of homoplasy… Distance methods Normally fast and simple e.g. UPGMA, Neighbour Joining, Minimum Evolution, Fitch-Margoliash Correction for multiple hits Only differences can be observed directly – not distances All distance methods rely (crucially) on this A great many models used for nucleotide sequences (e.g. JC, K2P, HKY, Rev, Maximum Likelihood) AA sequences are infinitely more complicated! Accuracy falls off drastically for highly divergent sequences Distance methods Attempts to account for multiple hits using models in distance methods (observed vs. estimated amount of evol. distance) Other Phylogenetic methods Maximum likelihood assumes a particular model of sequence evolution and calculates how likely each branch arose based on the character data Uses all data, even autapomorphies and invariant sites Uses models of evolution designed to capture a pattern of change across characters (e.g., DNA) Allows us to account for complex patterns of nucleotide evolution across regions of genes that may evolve very differently (thus, not all types of changes are weighted evenly in determining the phylogeny…) Lets look at an example… although we will save more heated discussions of patterns for Bayesian MCMCMC methods…. 1.8 2 Gene 1 1.8 1.6 Gene 2 C-G 1.4 1.2 A-C 1 0.8 0.6 0.4 0.2 285 240 195 Length Along Genome 150 105 60 15 504 441 378 315 252 189 126 F 63 00 C A-T 0 Relative Rate of Substitution (G-T = 1) Within vs. Between Gene Variation Transversions Maximum Likelihood Methods Likelihood methods are among the most accurate methods to reconstruct phylogenies! However, they are VERY VERY computationally intensive a tree with 30 species may take several days, with 100 species may take several months! New likelihood methods employing Bayesian statistics along with Marcov Chain Monte Carlo algorithms are helping to solve this problem and are the cutting edge of phylogeny reconstruction these days… Likelihood Methods Requires a model of evolution Each substitution has an associated likelihood given a branch of a certain length A function is derived to represent the likelihood of the data given the tree, branch-lengths and additional parameter So, the tree we get from ML is “the phylogeny that is most likely to have produced the observed data (under the model of evolution selected)” The Likelihood Criterion Given two trees, the one maximizing the probability of the observed data is best Site likelihood - probability of the data for one site conditional on the assumed model of evolution Tree score - sum of site log-likelihoods (term score also general term for the derivative of the lnL) Unlike parsimony tree lengths, log-likelihoods are comparable across models as well as trees Models can be made more parameter rich to increase their realism The most common additional parameters are: A correction to allow different substitution rates for each type of nucleotide change A correction for the proportion of sites which are unable to change A correction for variable site rates at those sites which can change The values of the additional parameters will be estimated in the process (e.g. PAUP) A gamma distribution can be used to model site rate heterogeneity Long Branches Attract In a set of sequences evolving at different rates the sequences evolving rapidly are drawn together Distance methods are VERY VERY prone to making this error Parsimony is also prone to this error Likelihood methods employ an ‘informed’ view of character change (a model) which helps identify situations which probably represent homoplasy, thus decreasing LBA Phylogenetic Methods… It is useful to use a variety of tree reconstruction methods If methods are congruent you have more confidence in your reconstructions! Reconstructing Phylogenies Phylogenies can be useful tools to answer important evolutionary questions One must always question the methods used to reconstruct the phylogeny to be confident in the results Comparison of methods Inconsistency Neighbour Joining (NJ) is very fast but depends on accurate estimates of distance. This is more difficult with very divergent data Parsimony suffers from Long Branch Attraction. This may be a particular problem for very divergent data NJ (and less so, MP) can suffer from Long Branch Attraction Parsimony is also computationally intensive Codon usage bias can be a problem for MP and NJ Maximum Likelihood is the most reliable but depends on the choice of model and is very slow Methods may be combined Finding the best tree…?? How do we find the best tree? With a small number of taxa you can evaluate all possible trees Exhaustive search With more taxa the amount of possible trees increase exponentially For 8 taxa in artiodactyl tree there are 10,395 possibilities Must use a shortcut method to evaluate trees Tree Space Search Strategies Exhaustive Branch and Bound Heuristic Branch Swapping Exhaustive Search Sequences Number of unrooted, binary trees 4 3 5 15 6 105 7 945 8 10,395 9 135,135 10 2,027,025 11 34,459,425 12 654,729,075 13 13,749,310,575 14 316,234,143,225 15 7,905,853,580,625 16 213,458,046,676,875 17 6,190,283,353,629,375 18 191,898,783,962,510,625 19 6,332,659,870,762,850,625 20 221,643,095,476,699,771,875 This run would just about be finished had we started it at the time prokaryotes diverged from eukaryotes (about 2.5 billion years ago!) Branch-and-bound At start, we know this tree has length 1982 steps Lineage out of contention (> 1982 steps) Lineage out of contention (> 1982 steps) (best) Ok, still under 1982 steps Theoretically predicted to always find the best tree -if using MP with a B&B search for 50 taxa, plan on between 2 days to 2 weeks Tree Islands This landscape has 5 peaks, only 1 of which represents the global optimum. best tree (globally optimal tree) locally optimal tree Imagine this depicts “tree space” Heuristic search algorithms are “hill climbers” – they only climb up -However, this type of search is typically the only choice we have for even small datasets… Solution Do 100-1000 replicates starting in different parts of tree space to find global optimum Heuristic search started here will not find global optimum How confident am I that my tree is correct? Bootstrap values Bootstrapping is a statistical technique that can use random resampling of data to determine sampling error for tree topologies How Reliable is a Phylogeny? How do we evaluate confidence in a tree? Bootstrap values are percentages of the number of times the same branch arose after repeated sampling Bootstrap support over 70% indicates that the correct relationship was probably found Investigators usually report any bootstrap value over 50% Bootstrapping phylogenies Characters are resampled with replacement to create many bootstrap replicate data sets Each bootstrap replicate data set is analysed (e.g. with parsimony, distance, ML etc.) Agreement among the resulting trees is summarized with a majority-rule consensus tree Frequencies of occurrence of groups, bootstrap proportions (BPs), are a measure of support for those groups Bootstrapping – How reliable is our phylogeny or part of our phylogeny?? Bootstrap replicate #1… …repeat this random resampling of date lots of times and see how many of these ‘pseudodatasets estimate a particular relationship.. Bootstrap - interpretation Bootstrapping is a very valuable and widely used technique (it is demanded by some journals) BPs give an idea of how likely a given branch would be to be unaffected if additional data, with the same distribution, became available BPs are not the same as confidence intervals. There is no simple relationship between bootstrap values and confidence intervals. There is no agreement about what constitutes a ‘good’ bootstrap value (> 70%, > 80%, > 85% ????) Some theoretical work indicates that BPs can be a conservative estimate of confidence intervals If the estimated tree is inconsistent all the bootstraps in the world won’t help you….. Bootstrap - interpretation Lets consider why bootstraps for a particular relationship may be low…. This can mean 2 things: There is conflicting signal in the data whereby some evidence supports one relationship while other evidence supports another Could be, simply, very little evidence overall (not conflicting evidence, but only one or two characters overall support this relationship) This is often the case if speciation is rapid and lineages split from one another (or a common ancestor) rapidly In general, rapid evolutionary radiations are nearly impossible to accurately estimate without enormous amounts of characters (because of low phylogenetic signal or evidence for exclusive common ancestry) Bootstrapping 59 71 Ochromonas Symbiodinium Prorocentrum Loxodes Tracheloraphis Spirostomumum Euplotes Tetrahymena Gruberia 16 59 26 71 16 21 Ochromonas Symbiodinium Prorocentrum Loxodes Spirostomumum Tetrahymena Euplotes Tracheloraphis Gruberia Majority-rule consensus (with minority components) Wim de Grave et al. Fiocruz bioinformatics training course MP – Tree length based tests of hypotheses Templeton test Compare the length (# steps) of the optimal tree vs. the length of tree that would result from the topological hypothesis you want to test Do this statistically using a 1-tailed Wilcoxan signed-rank test Likelihood-based tests of topologies Kishino-Hasegawa test Trees specified apriori KH can be used to test whether two competing hypotheses have significantly different likelihood NB should not be used to test trees that have been chosen on the basis of the data! Shimodaira-Hasegawa test Can be used to test confidence of ML tree compared to related trees (e.g. second most likely tree from the data)