Link to Powepoint - Computational Bioscience Program

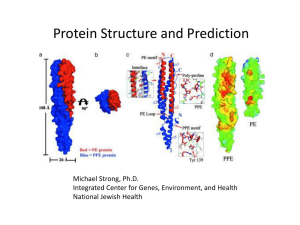
Protein Structure and Prediction
Michael Strong, Ph.D.
Integrated Center for Genes, Environment, and Health
National Jewish Health
Why do we care about protein structures
Structure leads to Function – Enzymes, Structural (Cell Wall),
Protein Interactions (host-pathogen), Replication, Transcription, Translation
Ebola Virus
-RNA genome
-Encodes 7 proteins
(Binds to receptors on a cell surface, attached carbohydrades help it evade immune system)
Shapes the virus, enables budding
Protects the RNA viral genome
To make new copies of RNA genome
Why do we care about protein structures
Combining Structure and Genomic Information
Help us understand phylogeny and implications of mutations
Science 12 September 2014: Vol. 345 no. 6202 pp. 1369-1372
Why do we care about protein structures
Combining Structure and Genomic Information
Help us understand phylogeny and implications of mutations
Drug Resistance
Tuberculosis
Rifampin rpoB drug target
Why do we care about protein structures
Combining Structure and Genomic Information
Help us understand phylogeny and implications of mutations
Homology model of CFTR structure, with common mutation F508 .
PNAS 3256–3261, 105:9
From Sequence to Structure
HIV Protease
PQITLWKRPLVTIRIGGQL
KEALLDTGADDTVLEEM
NLPGKWKPKMIGGIGGF
IKVRQYDQIPIEICGHKAI
GTVLVGPT
PVNIIGRNLLTQIGCTLNF
Experimental
Approach
X-ray Crystallography
NMR
Cryo-Electron Microscopy
HIV Protease
With Inhibitor
From Sequence to Structure
H1N1 NA
MNPNQKIITIGSVCMTIGMANLILQIG
NIISIWISHSIQLGNQN
QIETCNQSVITYENNTWVNQTYVNISN
TNFAAGQSVVSVKLAGNSSLCPVSGW
AIYSK
DNSVRIGSKGDVFVIREPFISCSPLECRT
FFLTQGALLNDKHSNGTIKDRSPYRTL
MS
CPIGEVPSPYNSRFESVAWSASACHDGI
NWLTIGISGPDNGAVAVLKYNGIITDTI
KS
WRNNILRTQESECACVNGSCFTVMTD
GPSNGQASYKIFRIEKGKIVKSVEMNAP
NYHY
EECSCYPDSSEITCVCRDNWHGSNRP
WVSFNQNLEYQIGYICSGIFGDNPRPN
DKTGS
CGPVSSNGANGVKGFSFKYGNGVWIG
RTKSISSRNGFEMIWDPNGWTGTDN
NFSIKQD
IVGINEWSGYSGSFVQHPELTGLDCIRP
CFWVELIRGRPKENTIWTSGSSISFCGV
NS DTVGWSWPDGAELPFTIDK"
Computational
Approach
Homology Modeling
Protein Threading
Ab initio
Protein Building Blocks
Typical Protein Sequence MNPNQKIITIGSVCMTIGMANLILQIGNIISIWISHSIQLGNQN
Protein Building Blocks
Amino Acid Side Chain (R groups)
Disulfide
Bonds
Amino Acid Side Chain (R groups)
Amino Acid Side Chain (R groups)
acidic
+ basic
Most Proteins Spontaneously Fold
DNA
Transcribed by RNA polymerase
RNA
Translated by Ribosome
Folded Protein
Some proteins need chaperones for correct folding
Most Proteins Spontaneously Fold
Folded protein
Christian
Anfinsen’s Experiment
1950s
Denaturing conditions
Unfolded protein
Native conditions native state, Folded protein spontaneous self-organisation
(~1 second)
Most Proteins Spontaneously Fold
Important to Computational Biologists, because this suggests that all information relating to the correct folding of a protein is contained in it’s primary amino acid sequence, but …
Most Proteins Spontaneously Fold
But Proteins lack easy rules for folding as compared to DNA
DNA
Protein
Many Factors Influence Protein Folding
Protein
Proteins Assume the Lowest Energy
Structure
Factors that influence folding include:
1. Hydrophobic Interactions / collapse
(particularly within the core)
2. Hydrogen bonds – lead to secondary structures
3. Disulfide Bonds (Cysteine residues)
4. Salt Bridges / Ionic Interactions
(among charged residues)
5. Multimeric interactions with same type or other proteins
Common Secondary Structures
Alpha helix
Common Secondary Structures
Beta Sheet
Common Secondary Structures
Loop Regions
Loop
Strong M et al, Proc National Academy of Sciences vol. 103 no. 21, 8060–8065, 2006
Example - Hemoglobin
Diversity of Protein Structures
B C D A
E
Rifampin target rpoB
Homology Model
Isoniazid Activating
Enzyme, KatG
Crystal Structure
F
Pyrazinamide
Activating enzyme pncA
Crystal Structure
G
Fluoroquinolone Target gyrA
Crystal Structure
H
Streptomycin resistance gidB
Homology model
Isoniazid Target inhA
Crystal Structure
Ethionamide
Target, inhA
Crystal Structure
Streptomycin
Resistance rpsL
Homology model
Experimental Methods of Structure Determination
X-ray crystallography
High resolution structure determination
Grow a protein Crystal
Experimental Methods of Structure Determination
X-ray crystallography
High resolution structure determination
Experimental Methods of Structure Determination
X-ray crystallography
High resolution structure determination
•Intensities and phases of all reflections are combined in a Fourier transform to provide maps of electron density
Phases determined by using heavy metals or selenomethionine (MAD)
Experimental Methods of Structure Determination
NMR – Nuclear Magnetic Resonance
High resolution structure determination
• Smaller Proteins than X-ray
• Distances between pairs of hydrogen atoms
• Lots of information about dynamics
• Requires soluble, non-aggregating material
• Assignment sometimes difficult
NOE cross-peak if they are within
5.0
Å
Experimental Methods of Structure Determination
Cryo Electron Microscopy
Low to medium resolution structure determination
• Low to medium resolution
~10-15Å
• Limited information about dynamics
• Can be used for very large molecules and complexes
Database of Protein Structures
PDB – Protein Data Bank
Database of Protein Structures
PDB – Protein Data Bank
104,125 structures as of 10/20/2014
Database of Protein Structures
PDB – Protein Data Bank
Even so, the number of solved structures greatly lags behind the rate of new genes being sequenced … Solution: Computational Structural Methods
GenBank Sequences
Database of Protein Structures
PDB – Protein Data Bank Files
• Atoms in pdb files are defined by their Cartesian coordinates:
Visualization of PDB files
Pymol, Jmol, Chimera, etc
Visualization of PDB files
Pymol, Jmol, Chimera, etc
DALI Structural Alignments
Align Protein Structures, Structure Superposition
Generates a comparison matrix (transform protein into a 2D array of distances between C-alpha atoms. Z score reflects reliability, lowest
RMSD identified
From Sequence to Structure
H1N1 NA
MNPNQKIITIGSVCMTIGMANLILQIG
NIISIWISHSIQLGNQN
QIETCNQSVITYENNTWVNQTYVNISN
TNFAAGQSVVSVKLAGNSSLCPVSGW
AIYSK
DNSVRIGSKGDVFVIREPFISCSPLECRT
FFLTQGALLNDKHSNGTIKDRSPYRTL
MS
CPIGEVPSPYNSRFESVAWSASACHDGI
NWLTIGISGPDNGAVAVLKYNGIITDTI
KS
WRNNILRTQESECACVNGSCFTVMTD
GPSNGQASYKIFRIEKGKIVKSVEMNAP
NYHY
EECSCYPDSSEITCVCRDNWHGSNRP
WVSFNQNLEYQIGYICSGIFGDNPRPN
DKTGS
CGPVSSNGANGVKGFSFKYGNGVWIG
RTKSISSRNGFEMIWDPNGWTGTDN
NFSIKQD
IVGINEWSGYSGSFVQHPELTGLDCIRP
CFWVELIRGRPKENTIWTSGSSISFCGV
NS DTVGWSWPDGAELPFTIDK"
Secondary Structure Prediction
Alpha Helix, Beta Strand, or Other
Computational
Approach
Tertiary Predictions:
1. Homology Modeling
2. Fold Recognition
3. De Novo Protein Structure
Prediction
Secondary Structure Prediction
-probability of amino acid to be in a alpha helix, Beta strand, or other (coil/loop) based on known structures.
-Chou-Fasman (short runs of amino acids), GOR (Bayesian, takes neighbors into account)
- helices – no prolines, periodicity 3.6 residues/turn
- strands – alternating hydropathy, or ends hydrophillic and center hydrophobic
-other – small, polar, flexible residues, and prolines
But, stalled at 55- 60% accuracy
Also used position specific profiles based on multiple sequence alignments (evolutionary information) (ie insertion/deletion more likely to be in coil/turn), PSI BLAST and HMM, NN and SVM
(improved to about 75-80%)
Secondary Structure Prediction
But we really want to know how the protein folds in three dimensions
CASP - Critical Assessment of Techniques for
Protein Structure Prediction
• Started in 1994, Helped push the field of structure prediction
•“Contest-like” setup
•Catagories include:
•Homology Modeling / Comparative Modeling
•Fold Recognition / Threading
•Ab Initio, De novo
•Partially vs. Automated Methods (now quite similar results)
Goal: Predict structures of solved but unpublished/unreleased structures (used to evaluate predictions. Every year, predictions / algorithms get better
Comparative Modeling “Homology Modeling”
• Proteins that have similar sequences (i.e., related by evolution) are likely to have similar three-dimensional structures
1. BLAST sequence of Interest against PDB to identify a template
•Multiple templates can be used if desired
•Templates with Ligands bound can be used to identify binding sites and interacting residues in the homology model
Sequence identity required depends on protein length. A good rule of thumb is to have at least 40% sequence identity . Higher sequence identity is best. Lower than 25% is not reliable (zone of uncertainty)
Above 75% sequence identity, usually quite reliable homology model
Accurate sequence alignments very important
Programs include Modeller and Swiss Model
Comparative Modeling “Homology Modeling”
Steps include:
1. Template recognition and initial alignment
2. Alignment Correction (Multiple Sequence Alignment can
Help)
3. Backbone Generation (transfer coordinates from template)
4. Loop Modeling (loops hard to predict with insertions)
5. Side Chain Modeling (usually similar tortion angles at high sequenc ID)
6. Model Optimization (minor energy minimization steps or restrain some atom positions)
7. Model Validation (Higher ID more accurate usually,
Calculate energy, or normality index (bond length, tortion angles))
8. Iteration (to refine)
Protein Threading, Fold Recognition
Often, seemingly unrelated proteins adopt similar folds.
-Divergent evolution, convergent evolution. For sequences with low or no sequence homology
Protein Threading
§ Generalization of homology modeling method
• Homology Modeling: Align sequence to sequence
• Threading: Align sequence to structure (templates)
For each alignment, the probability that that each amino acid residue would occur in such an environment is calculated based on observed preferences in determined structures.
§ Rationale:
• Limited number of basic folds found in nature
• Amino acid preferences for different structural environments provides sufficient information to choose the best-fitting protein
fold (structure)
Fold recognition
• The number of possible protein structures/folds is limited (large number of sequences but relatively few folds ( some estimate ~1000 )
)
(most apparent when 50% of structures with no seq homology were solved and had folds similar to known structures)
90% of new structures deposited in PDB have similar folds to those already known
• Proteins that do not have similar sequences sometimes have similar threedimensional structures (such as B-barrel TIM fold)
3.6 Å
5% ID
NK-lysin (1nkl) Bacteriocin T102/as48 (1e68)
• A sequence whose structure is not known is fitted directly (or “threaded”) onto a known structure and the “goodness of fit” is evaluated using a discriminatory function
• Need ways to move model closer to the native structure
Ab initio prediction of protein structure – concept
Difficult because search space is huge . Much larger conformational space
Goal: Predict Structure only given its amino acid sequence
In theory: Lowest Energy Conformation
• Go from sequence to structure by sampling the conformational space in a reasonable manner and select a native-like conformation using a good discrimination function
Difficult for sequences larger that 150aa
Rosetta (David Baker lab) one of best (CASP evaluation)
Rosetta structure prediction
2 phases
1. Low-resolution phase – statistical scoring function and fragment assembly
A. local structure conformations using info from PDB (3 and 9mer stretches)
B. multiple fragment substitution simulated annealing – to find best arrangement of the fragments (Monte Carlo
Search)
C. low resolution ensemble of decoy conformations
2. Atomic refinement phase using rotamers and small backbone angle moves (in populated regions of Ramachandran plot)
A. Refinement
B. Then structures clustered based on RMSD
C. Center of the Largest Clusters chosen as representative folds (likely to be correct fold)
Quality Assessment
Ramachandran Plot – Phi Psi angles
To identify residues that may be in wrong conformation
Procheck, What_check