Personalized Medicine of Deafness
advertisement

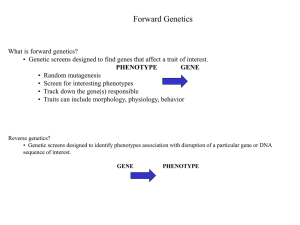
Personalized Medicine of Deafness Marci M. Lesperance, MD, FACS, FAAP Professor and Division Chief, Pediatric Otolaryngology O T O LAR Y N G O LO G Y - H EAD & N E C K S U R G E R Y University of Michigan Health System CANC deafness diabetes CV disease Head/neck breast adrenal colon Types of deafness • Conductive • Sensorineural • Auditory neuropathy • Syndromic vs. nonsyndromic Hearing Loss • Most common sensory disorder, affects 28 million Americans • Over 500 syndromes such as Usher (deafness and retinitis pigmentosa causing blindness) • 50% develop presbycusis (age-related HL) by age 80 • 2 in 1000 children are born with educationally significant hearing loss; 5 in 1000 by adolescence • Association studies of presbycusis show increased risk with smoking, high BMI, noise exposure; moderate alcohol consumption is protective • SNHL can be rehabilitated but not cured The inner ear • • • • Inaccessible, within the temporal bone Tiny quantities of proteins Genetics Animal models – Proteomics – Histopathology Richardson et al, Ann Rev Physiol 2011 Positional cloning/ positional candidate approach to discovering novel genes Human Genome Working Draft GENE1 GENE2 GENE3 …… GENE200 Dror and Avraham, Neuron 2010 DFNA2 DFNA37 DFNB32 DFNA7 DFNA49 DFNB36 DFNA34 DFNB45 DFNB7/11 DFNA47 DFNA51 DFNA36 DFNB31 DFNB33 DFNB3 DFNA20/26 2010 2012 DFNA43 DFNB9 DFNB47 DFNB58 DFNB27 DFNA16 DFNB59 DFNB30 DFNB23 DFNA19 DFNB12 DFNB57 DFNB19 DFNA6/14 DFNB6 DFNB42 DFNA18 DFNB15 DFNA44 DFNB51 DFNA32 DFNB18 DFNB63 DFNB2 DFNA11 DFNA8/12 DFNB21 DFNB24 DFNB20 DFNB68 DFNA57 DFNB15 DFNA4 DFNA38 DFNB25 DFNB55 DFNA27 DFNA24 DFNB26 DFNA39 DFNA42 DFNA52 DFNB49 DFNB60 DFNA1 DFNA15 DFNA42 DFNB66 DFNA13 DFNB53 DFNA22 DFNB37 DFNB31 DFNA21 DFNB38 DFNA10 DFNB44 DFNA5 DFNB39 DFNB4 DFNB14 DFNB17 DFNB13 DFNA28 DFNA50 DFNB62 DFNA31 DFNA48 DFNA25 DFNA41 DFNB50 DFNB1 DFNA3 AUNA1 DFNA9 DFNB5 DFNA23 DFNB35 DFNA53 DFNB16 DFNA30 DFNB48 DFNA40 DFNB22 DFN6 DFN4 DFNB65 DFNB8/10 DFNB29 DFNA17 DFNB28 DFNB40 DFN3 DFN2 DFNY Courtesy Heidi Rehm, PhD Connexin 26 (GJB2), DFNB1 • Mutations account for 15-50% of recessive non-syndromic SNHL in US population • Testing widely available ($300-$500) • One mutation (35delG) accounts for 75-80% of all mutations in Caucasians • 35delG homozygotes typically have congenital profound SNHL, but modifiers likely exist • 3.01% carrier rate of 35delG in Iowa population (Green et al. 1999) 35delG/+ +/+ Patterns of sensorineural hearing loss Frequency in Hertz (Hz) 250 500 750 1000 1500 2000 0 X O Hearing Threshold (dB) 10 20 30 X 40 O 50 O X O X X 3000 4000 O X Frequency in Hertz (Hz) 6000 8000 -10 O X O X 60 70 80 0 10 Hearing Threshold (dB) -10 20 X 500 750 1000 1500 2000 3000 4000 ][ O X O X O 30 O X 40 [ ][ X O 50 O X 6000 8000 O X O X O X 60 70 90 80 100 90 Low frequency SNHL WFS1 mutations Also DIAPH1, FGFR3 250 O [ X ] Frequency (Hz) vs Right A/C Frequency (Hz) vs Right B/C Frequency (Hz) vs Left A/C Frequency (Hz) vs Left B/C 100 Mid frequency “cookie-bite” TECTA (alpha tectorin) mutations High frequency or flat audiogram: many genes Genetic testing for nonsyndromic auditory neuropathy • OTOF (otoferlin) – 48 coding exons and splice sites; recessive – $1500 through Partners- does not accept 3rd party payment • Pejvakin – reported in two families from Iran • DIAPH3 (Schoen et al. 2010, PNAS) – Autosomal dominant auditory neuropathy; currently no lab offering clinical testing BCN view of the world • Does a definitive dx remain uncertain even after H&P exam and conventional diagnostic studies? • Does the patient display clinical features related to the inherited mutation in question? • What justifies the patient’s increased risk? i.e. family history, ethnic background • Is this disease treatable or preventable? • Will the result of the test directly influence the tx being delivered to the patient? • Requires informed consent and genetic counseling Deafness gene platforms • OtoChip™- 19 assorted genes for deafness including Usher, mitochondrial, $3800 • Otogenetics: 80+ genes, $488/sample • OtoSCOPE: 60+ genes, $2000/sample • Missing most “syndromic” genes such as: – FGF3 : deafness, microtia, and microdontia (DFNB63) – BSND: Bartter syndrome, DFNB73 (mild renal dysfunction) – FGFR3: Muenke craniosynostosis; CATSHL (camptodactyly and tall stature with HL) Whole exome sequencing • All coding exons and intron-exon junctions • At least 2 samples/family to reduce number of variants for follow-up • Cross referencing to linkage region(s) • Trending toward whole genome sequencing (at some lower coverage) – Unknown exons expressed specifically in the ear – Splice mutations, distant enhancers/promoters – Better sensitivity for deletions/duplications Annotation • dB SNP filter – Includes deafness mutations such as 35delG – Allow minor allele frequency <5% • Exome variant server (U Washington) filter • Up to 10,000 alleles • Subjects not necessarily screened for deafness; some reported deafness mutations seen • Splice mutations may be called as non-synonomous or synonomous variants • Deafness mutations may occur in noncoding sequence (5’UTR mutation in DIAPH3) Interpretation • Pooling samples reduces costs but may sacrifice coverage of particular linkage region • Failure to confirm on Sanger sequencing • Easier to detect homozygous mutations causing recessive disease • Dominant requires analysis of every variant and confirmation of segregation in the family Most common non-genetic cause: Human cytomegalovirus infection • • • • HCMV affects 1.1/1000 live births 5% symptomatic; 30-65% incidence of SNHL 95% are asymptomatic; 8-15% incidence SNHL Must detect in first 3 weeks of life; universal screening of blood or saliva; newborn blood spots have low sensitivity (JAMA 2010) • Antibody testing may be able to exclude HCMV as an etiology The future is here! • Identifying a genetic etiology helps with prognosis and recurrence risk • Economies of scale will bring the price down • Developing a clinical algorithm and drawing genotypephenotype correlations Interval Genomic Viewer plots 3 SNPs in ZNF575 Average 10x chr19:44039571 chr19:44039572 chr19:44039574 Alignment error PCR duplication artifact Note poor coverage of 3rd sample Jishu Xu 1 SNP in KCNN4 chr19:44271753 A19:4T (23x) A34:6T (40x) AA 14:0 (14x) PCR duplication artifact Utility of chromosomal microarray analysis (CMA) for evaluation of deafness • • • • • GJB2 null alleles due to distant deletions ~100kb deletion of STRC (DFNB16) CHD7 (CHARGE syndrome) TWIST (Saethre-Chotzen syndrome) Recessive disorders with 0-1 mutations (e.g., SLC26A4 gene ) 555kb deletion of chromosome 16p13.3 Non-syndromic Sensorineural Hearing Loss: Summary • Recessive is usually congenital and profound; GJB2 (also pseudodominant) • Dominant is usually delayed onset and progressive – Low frequency WFS1 – Mid frequency “cookie-bite” TECTA (alpha-tectorin) • Auditory neuropathy: OTOF, DIAPH3 • Otherwise, no predominant genes; some in single families or yet unidentified • Next generation sequencing technologies Genotype cohorts provide some prognostic information Snoeckx et al. GJB2 mutations and hearing loss: a multicenter study, Am J Hum Genet, 2005. OtoChipTM • 19 genes, 70,000 bases; $3800, 8 week TAT • Nonsyndromic SNHL: CDH23, DFNB31 (WHRN), GJB6, MYO6, MYO7A, OTOF, PCDH15, SLC26A4 (PDS), TMC1, TMIE, TMPRSS3, USH1C • Mitochondrial tRNAser(UCN) and 6 in 12S rRNA • OTOF, SLC26A4 • Usher syndrome : CDH23, CLRN1, DFNB31, GPR98 (exons 8, 20, 31-41 & 89), MYO7A, PCDH15, USH1C, USH1G, USH2A • Not included : WFS1, TECTA, KCNQ4 OtoSCOPE (Shearer et al. 2010) • 50 nonsyndromic deafness genes, including mitochondrial and miRNA, and 4 Usher genes • Identified genetic etiology in 5/6 unknowns – p.A366T/p. N1098S in CDH23 (DFNB12, USH1D) – ~100kb deletion in STRC/p.Q1353X – p.D288DfsX17 in MYO6 (DFNA22) – p.E1965X in MYH14 (DFNA4) – p.L281S in KCNQ4 (DFNA2) – Found 4 variants in one dominant case, none segregating with deafness- etiology unknown AudioGene http://audiogene.eng.uiowa.edu • Input the audiometric thresholds and AudioGene will predict the most likely genes for dominant SNHL KCNQ4 • Not available for all genes • Can’t analyze more than one audiogram at a time • Can view audioprofiles for selected genes • Often, noise exposure alters the classic pattern High frequency SNHL • • • • KCNQ4 mutation Noise exposure Presbycusis Or many other genes! 1 SNP in SARS2 chr19:39408746