ppt
advertisement

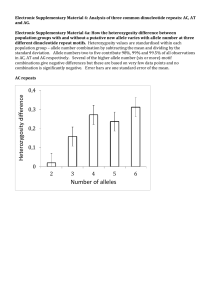
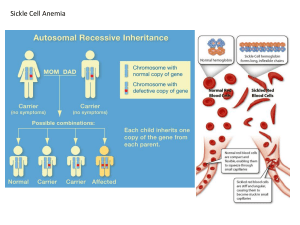
Lecture 5: Genetic Variation and Inbreeding January 24, 2014 Last Time Hardy-Weinberg Equilibrium Using Hardy-Weinberg: Estimating allele frequencies for dominant loci Variance of allele frequencies for dominant loci Hypothesis testing Today Measures of diversity More Hardy-Weinberg Calculations Merle Patterning in Dogs First Violation of Hardy-Weinberg assumptions: Random Mating Effects of Inbreeding on allele frequencies, genotype frequencies, and heterozygosity Expected Heterozygosity If a population is in Hardy-Weinberg Equilibrium, the probability of sampling a heterozygous individual at a particular locus is the Expected Heterozygosity: 2pq for 2-allele, 1 locus system OR 1-(p2 + q2) or 1-Σ(expected homozygosity) n HE 1 more general: what’s left over after calculating expected homozygosity 2 p i, i 1 Homozygosity is overestimated at small sample sizes. Must apply correction factor: Correction for bias in parameter estimates by small sample size HE n 2 1 p i , 2N 1 i 1 2N Maximum Expected Heterozygosity Expected heterozygosity is maximized when all allele frequencies are equal Approaches 1 when number of alleles = number of chromosomes 2N H E (max) 1 i 1 2 2N 1 1 1 1 2N 2N 2N 2N Applying small sample correction factor: HE 2 n 2N 2N 1 2 1 p i 1 2N 1 i 1 2N 1 2N 2N Also see Example 2.11 in Hedrick text Observed Heterozygosity Proportion of individuals in a population that are heterozygous for a particular locus: HO N N ij H ij Where Nij is the number of diploid individuals with genotype AiAj, and i ≠ j, And Hij is frequency of heterozygotes with those alleles Difference between observed and expected heterozygosity will become very important soon This is NOT how we test for departures from HardyWeinberg equilibrium! Alleles per Locus Na: Number of alleles per locus Ne: Effective number of alleles per locus Same as ne in your text If all alleles occurred at equal frequencies, this is the number of alleles that would result in the same expected heterozygosity as that observed in the population Ne 1 , Na i 1 2 pi Example: Assay two microsatellite loci for WVU football team (N=50) Calculate He, Na and Ne Locus A Locus B Allele Frequency Allele Frequency A1 0.01 B1 0.3 A2 0.01 B2 0.3 A3 0.98 B3 0.4 HE n 2 1 p i , 2N 1 i 1 2N Ne 1 , Na i 1 p 2 i Measures of Diversity are a Function of Populations and Locus Characteristics Assuming you assay the same samples, order the following markers by increasing average expected values of Ne and HE: RAPD SSR Allozyme Example: Merle patterning in dogs Merle or “dilute” coat color is a desired trait in collies, shetland sheepdogs (pictured), Dachshunds and other breeds Homozygotes for mutant gene lack most coat color and have numerous defects (blindness, deafness) Caused by a retrotransposon insertion in the SILV gene Clarke et al. 2006 PNAS 103:1376 Example: Merling Pattern in collies Homozygous wild-type M1M1 N=6,498 Heterozygotes Homozygous mutants M1M2 M2M2 N=3,500 N=2 Is the Merle coat color mutation dominant, semi-dominant (incompletely dominant), or recessive? Do the Merle genotype frequencies differ from those expected under Hardy-Weinberg Equilibrium? Why does the merle coat coloration occur in some breeds but not others? How did we end up with so many dog breeds anyway? Nonrandom Mating: Inbreeding Inbreeding: Nonrandom mating within populations resulting in greater than expected mating between relatives Assumptions (for this lecture): No selection, gene flow, mutation, or genetic drift Inbreeding very common in plants and some insects Pathological results of inbreeding in animal populations Recessive human diseases Endangered species http://i36.photobucket.com/albums/e4/doooosh/microcephaly.jpg Important Points about Inbreeding Inbreeding affects ALL LOCI in genome Inbreeding results in a REDUCTION OF HETEROZYGOSITY in the population Inbreeding BY ITSELF changes only genotype frequencies, NOT ALLELE FREQUENCIES and therefore has NO EFFECT on overall genetic diversity within populations Inbreeding equilibrium occurs when there is a balance between the creation (through outcrossing) and loss of heterozygotes in each generation Inbreeding can be quantified by probability (f) an individual contains two alleles that are Identical by Descent P A1A2 F1 A3A4 A1A3 F2 A 3A 3 A 2A 3 A2A3 Identical by descent (IBD) A 1A 2 A3A4 A1A3 A2A3 A3A5 A3A3 A2A3 Identical by state (IBS) Identical by descent (IBD) Nomenclature D=X=P: frequency of AA or A1A1 genotype R=Z=Q: frequency of aa or A2A2 genotype H=Y: frequency of Aa or A1A2 genotype p is frequency of the A or A1 allele q is frequency of the a or A2 allele All of these should have circumflex or hat when they are estimates: pˆ Effect of Inbreeding on Genotype Frequencies D fp p (1 f ) 2 D fp p fp 2 D p fp fp 2 2 2 D p fp (1 p ) 2 D p fpq 2 R q fpq 2 H 2 pq 2 fpq fp is probability of getting two A1 alleles IBD in an individual p2(1-f) is probability of getting two A1 alleles IBS in an individual Inbreeding increases homozygosity and decreases heterozygosity by equal amounts each generation Complete inbreeding eliminates heterozygotes entirely Fixation Index The difference between observed and expected heterozygosity is a convenient measure of departures from Hardy-Weinberg Equilibrium F HE HO HE 1 HO HE Where HO is observed heterozygosity and HE is expected heterozygosity (2pq under Hardy-Weinberg Equilibrium)