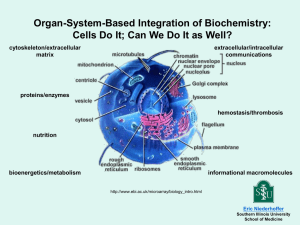
Powerpoint - Department of Agronomy and Plant Genetics
advertisement

Developing new SSR markers for barley derived from the EST database Karen A. Beaubien and Kevin P. Smith* Dept of Agronomy & Plant Genetics, University of Minnesota, St Paul, MN, *Corresponding Author: PH: (612) 624-1211; E-mail: smith376@umn.edu INTRODUCTION Genetic mapping of germplasm relevant to plant breeding programs with molecular markers serves to connect the wide array of valuable germplasm and genetic variation used in agriculture to the expanding frontier of plant genomics. Progress in mapping genes for important traits in barley and utilizing markers to improve the efficiency of breeding is dependent on the availability, cost, and ease of use of molecular markers. There are currently 632 SSR marker primer sets that have been developed of which 242 have been mapped in barley (Saghai Maroof et al., 1994; Liu et al., 1996; Ramsay et al., 2000). More recently, several studies have reported the development of an additional 202 SSR markers (Li et al., 2003, Thiel et al., 2003), however, most of these markers mapped to regions of the barley genome that already have good coverage with SSR markers (Table 1). There is still a need for additional publicly available SSR markers to improve coverage of the barley genome and facilitate breeding and genetics research. 0 3 6 8 9 10 11 12 15 17 18 21 22 23 24 26 27 30 33 35 38 40 41 42 43 48 53 58 61 65 66 67 70 71 72 74 120 122 123 ABG704 MWG036B RISIC10A Bmag0767 BCD129 MWG851A MWG555A ABG320 HVM004 iEst5 His3A Bmag0206 WG789A MWG089 MWG530 ABC167A dRcs1 ABG380 Mad1 MWG564 ABC158 ABC151A KSUA1A ABC154A MWG836 UMB106 UMB107 Brz ABC156D BCD098 MWG649A HVM014b MWG911B BCD147 ABC308 ABG476 BCD349 ABC154B UMB104 Bmag0110a KFP194 Bmag0321 Amy2 ABC255 Bmac0187 Bmac0064 Bmag0482b ABG156B Ubi1 MWG571D TLM1 ABC310B KFP190 RISP103 bBE54E ABG608 ABC305 UMB105 Bmac0156 ABG497B ABG461A UMB101 UMB102 WG420 RZ682 ABG652A ABC253 135 137 138 139 140 HVM005 HVPRP1B MWG635B UMB103 PSR106B 76 79 80 81 82 88 93 94 95 98 100 104 105 106 107 111 114 118 RESULTS Developed and evaluated 4 methods for designing SSR markers which differed in efficiency (Table 2). 238 different primer pairs were designed and screened. 44 new markers developed that produce 2-6 alleles among 12 mapping parents (Figure 2). 31 markers mapped using the Chevron x M69, Fredrickson x Stander, and Steptoe x Morex mapping populations (Figure 1). Markers are distributed throughout the barley genome. Most do not map to the target region for which they were designed. 16 of the 31 mapped markers map to BINs identified as having poor coverage with currently available SSR markers. 0 5 7 9 Integrate new EST-derived barley SSR markers on a barley consensus map derived from three mapping populations. ABG058 ABG703B CDO057A 16 SC1373 22 25 27 28 29 30 31 33 36 40 43 44 45 49 51 ABG008 UMB205a RbcS UMB203 BCD351F ABG318 ABG002 ABC156A HVM036 UMB205b MWG858 ABG358 ABC311 ABG459 Bmag0174c Pox ABG005MWG520A Adh8 ABG156A Bmac0218b cMWG663A UMB202 MWG887A MWG557 Bmac0140ABG316C UMB201 Bmac0093 EBmac0557 HVBKASI ABC167B Bmac0129b EBmac0558 EBmac0521b GMS003 HVM023 UMB204 GBM1024 EBmac0521c EBmac0521a ABC306 EBmac0615 bBE54D Bmag0140 Bmag0378 CDO588ABG014 BCD1087 TLM3 ABC256 ABG619MWG950 His3C ABC152DBmag0003c Bmag0003d MWG865 Rrn5S1 Vrs1 KSUF15 Bmag0125 MWG503 GBM1062 MWG503B Bmac0144g MWG882B MWG503A Bmac0144b Crg3A ABG072 ABG497C MWG649C STS3B-142 ABC252 EBmac0415 HVM054 ABC157 ABC153 ABG317B ABG317A ABG010B ABG316E ABG010CPcr1 cMWG720 BG123A bBE54C 67 68 69 70 71 72 73 74 76 77 79 80 83 85 86 89 90 91 92 94 95 99 102 105 106 109 111 112 117 123 124 126 128 131 136 137 139 140 141 145 150 153 155 165 0 1 2 3 4 6 13 15 17 23 26 30 34 43 44 45 47 49 50 51 ASE1A MWG878A 57 58 60 62 64 65 66 GBM1036 Develop PCR primers flanking SSRs in barley ESTs and screen for polymorphisms on a set of barley mapping parents. We evaluated four methods of designing EST-SSR markers. Methods 1, 2, and 3 were based on identifying syntenous regions in barley using rice genome sequence. Method 4 used expression specific ESTs. 3 (3H) 13 52 MATERIALS AND METHODS Increase the number of available SSR markers in BINs with poor coverage (Table 1). 2 (2H) 1 (7H) OBJECTIVES 52 53 54 55 56 58 61 63 67 68 70 72 75 76 77 78 81 82 85 89 95 100 105 117 120 122 124 125 127 128 132 136 140 142 144 146 149 153 161 4 (4H) ABA307B MWG571C STS3B-94 STS3B-47 UMB301 STS3B-65 STS3B-80 STS3B-52 ABC171 CDO395 MWG798B MWG584 ABG471 ABC171A Bmag0828 UMB302 Bmag0138 UMB304 Bmag0010c UMB303 Bmag0482a HVDHN7 BCD828 HVM009 Bmag0131 ABG399A Bmag0122 Bmac0067 Bmac0129a Bmag0136 Bmag0905 Bmag0603 Bmag0006 Bmag0482c MWG680 Bmag0131 Bmag0828 PSR156A CDO105C MWG571B MWG2227C ABG377 Bmag0225 MWG555B HVM060 ABG315 Bmag0010b ABG453 PSR78 MWG571A CDO345 ABG499 HVM070 CDO113B Bmag0606 His4B Bmag0482d ABG004 MWG803 WG110 ABC151C mPub BCD147B Bmag0013 ABC161 Bmag0877 MWG902 ABG654A Glb4 iBgl ABG495B EBmac0541 MWG838 ABG319B ABC172 Figure 2. Sample gel images from 4 markers. Markers are screened on 12 mapping parents (from left): Atahualpa; M81; Chevron; M69; Fredrickson; Stander; Harrington; OUH602; Hor211; Lacey; Steptoe; Morex. Methods 1-3: Sequenced markers (RFLP and STS) from barley BINs with poor SSR coverage were BLASTed against all Rice BAC sequences. Contiguous BAC sequences with multiple strong hits to markers from the same barley BIN were BLASTed against all barley ESTs with the low complexity filter turned off. The resulting barley ESTs were processed through the Tandem Repeats Finder (TRF) (Benson, 1999) to locate SSR motifs (1-6 bases in the repeat unit). Primer pairs were designed to flank SSR motifs using Primer3 software. All three methods used the same steps, but differed in the level of homology between the rice BAC and barley EST and the degree to which the SSR region in the EST matched the pattern of a perfect repeat unit. 0 1 12 15 18 20 24 32 33 35 37 41 45 48 50 51 52 53 54 55 56 57 58 59 60 61 63 64 67 68 70 73 74 77 82 85 88 90 94 96 97 98 100 101 103 106 118 120 123 124 136 137 140 147 ABG313B MWG077 CDO669A HVM040 B32E BCD402B BCD351D ABG705C MWG635A UMB401 TubA1 BCD1087 Dhn6 ABG715 WG1026B ABC303 ABA003 Adh4 ABG484 BCD1087a ABC321 STS3B142 MWG058 Bmac0181 KFP195 EBmac0906 HVM003 Bmag0808 Bmag0740 WG464 Bmag0306 bBE54A HVM068 MWG632D Bmac0310 BCD453B EBmac0775 ABG472 ABG319A EBmac0635 EBmac0701 EBmac0788 CDO020 iHxk2 ABG498 ABG500B HVMLOH1A bAP91 ABG366 ABG397 ABG319C Bmag0138b ABC305B HVM067 Bmy1 ksuH11 ASE1C ABA307A 0 3 6 11 12 16 18 19 25 30 33 34 38 39 42 44 45 46 47 48 50 51 53 55 57 58 60 61 64 65 68 69 76 79 80 87 90 92 95 96 98 107 113 117 123 125 130 132 134 136 139 141 Method 4 (Expression specific) One-hundred –seventy-three barley ESTs were identified from a Fusarium-infected library that do not have homology to any other publicly available barley and wheat ESTs in Genbank (as of April 9, 2003). These Fusarium-specific barley ESTs were processed through TRF to locate SSR motifs. Primer pairs were designed to flank SSR motifs with 60-100% matches to the repeat unit using Primer3. 7 (5H) 6 (6H) 5 (1H) MWG634 WG622 •Method 1 (Stringent) Barley ESTs processed through TRF had e-value between 0 and 1e-35. Primers were designed for SSRs with 85-100% matches to the repeat unit. •Method 2 (Relaxed) Barley ESTs processed through TRF had e-value between 0 and 1e-100. Primers were designed for SSRs with 50-100% matches to the repeat unit. •Method 3 (BLAST repeats) Isolated small repeats in the Rice BACs using TRF and BLASTed only the portion of the BAC that contained the SSR motif and some flanking sequence to the barley ESTs. Barley ESTs processed through TRF had e-value between 1e-150 and 1e-2. Primers were designed for SSRs with 85100% matches to the repeat unit. UMB502 MWG835A MWG938 MWG036A ACT008 MWG837 Hor1 Bmac0144a MWG2077 ABA004 MWG801B SC4-5B-1 BCD098 BCD738 ABG053 HVM043 Bmac0032 Ica1 MWG2040D ABC706C MWG2040C Bmac0144i ABG500A CDO580 UMB501b UMB501a ABC152B Bmac0144h ABC164A Bmag0770 WG789B HVM020 Bmac0090 ABR337 ABG074 Glb1 Bmag0872 ABC160 ABG452 BCD098 Bmac0213 ABG464 ABC156B ABC165C His3B ABC307A cMWG706A ABC257 cMWG733A AtpbA ABG702A ABC322B ABC261 Cab2 Aga7 MWG912 ABG387A 0 2 ASE1B ABG062 0 Act8D MWG620 Nar1 7 Act8B 14 ABG378 16 MWG502 23 ABC152A 36 46 47 48 49 51 52 54 55 56 60 61 63 64 73 77 78 86 87 90 93 97 UMB704 Bmac0163 Bmac0306 EBmac0970 Bmag0323 UMB705 Bmag0337 CDO348D MWG635G MWG635F MWG635D ABG497A MWG635H MWG2227B MWG635E ABC302A MWG503B ABG497B ABC168 ABC717 Bmag0812 UMB706 Bmag0113 UMB701 7 9 30 31 34 38 44 47 49 52 56 57 59 60 62 63 64 65 66 68 69 71 72 73 74 75 76 78 84 86 90 93 99 107 115 118 120 cMWG652A MWG2040E MWG2218 Bmag0500 PSR106 ABG387B MWG916 Bmg0001 ABG458 MWG887B Bmag0807 ABC169B CDO497 Bmag0173 Bmac0018 Bmag0003e Bmag0870 BCD340E Bmag0003a MWG2227A ksuD17 Bmac0218a Bmag0613 ABC175 EBmac0806a MWG820 EBmac0602 Bmac0251 EBmac0806b Nar7 Amy1 bBE54B MWG934 ABC170A ABG461B UMB602 Bmac0040 MWG549A 131 134 137 138 140 ABG713 MWG514 UMB603 UMB601 MWG798A MWG2196 149 cMWG684A S/M C/M & F/S Consensus 0 2 7 9 11 Mad2 MWG502 ABC483 MWG920A ABG316B 39 44 47 48 51 54 57 60 62 64 68 73 77 ABG705A ABG708 ABG395 CDO749 Ubi2 ksuA3A WG541 WG530 UMB707 WG889 ABC324 UMB703 ABC302A 89 CDO57B 95 mSrh 100 WG364 0 CDO400B 15 CDO059B 25 28 33 37 40 43 44 UMB702 MWG2227B ABG010D GMS061 EBmatc0003 ABG495A GMS027 134 139 143 145 149 152 MWG514B CDO504 iEst9 WG908 MWG877 ABG495A 54 ABG496 159 ABC155 166 ABC482 172 174 ABG391 ABG390 181 183 187 190 194 CDO484 ABG463 ABG314B MWG851C MWG851B 67 ABG391 73 ABG392 89 90 MWG851B MWG632C GMS001 Figure 1. Barley consensus maps developed from Chevron x M69 (C/M), Fredrickson x Stander (F/S), and Steptoe x Morex (S/M) mapping populations. Previously available SSR markers are in Maroon. New UMB SSR markers are in Blue. Maps and marker information are available at http://agronomy.coafes.umn.edu/barley. UMB704 CONCLUSIONS Method 3 was the most efficient method of producing useful SSR markers. UMB104 Approximately half of the barley genome has good coverage with SSR markers. UMB603 UMB702 Table 1. SSR marker coverage in barley by chromosome. Chromosome 1 Bins with good SSR coverage 1--2; 5--13 Bins with poor SSR coverage 3--4 2 3 4 5 6 4--12 6--8 6--9 5--7 6--8 1--3;13--15 1--4;8--16 1--4;10--13 1--4;9--14 1--5;9--14 7 3--5 1--2;6--15 Table 2. Comparison of four methods for designing EST based SSR markers. Method 1 2 3 Pairs tried 44 51 79 4 Overall 64 238 Produced Product Polymorphism Number % Number % 27 61.4 6 13.6 15 29.4 3 5.9 51 64.5 35 44.3 15 108 23.4 45.3 0 44 0.0 18.5 New methods must be developed to develop markers for underrepresented regions of the genome. ACKNOWLEDGEMENTS This research was supported by the North American Barley Genome Project. Special thanks to Charles Gustus for help in constructing the linkage maps. REFERENCES Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research 27: 573580. Li , J.Z., T. G. Sjakste, M. S. Röder, M. W. Ganal. 2003. Development and genetic mapping of 127 new microsatellite markers in barley. Theor. Appl. Genet. 107:1021–1027. Liu, Z.-W., R. M. Biyashev and M. A. Saghai Maroof, 1996 Development of simple sequence repeat markers and their integration into a barley linkage map. Theor. Appl. Genet. 93: 869–876. Ramsay, L., M. Macaulay, S. degli Ivanissevich, K. MacLean, L. Cardle, J. Fuller, K. J. Edwards, S. Tuvesson, M. Morgante, A. Massari, E. Maestri, N. Marmiroli, T. Sjakste, M. Ganal, W. Powell, and R. Waugh. 2000. A Simple Sequence Repeat-Based Linkage Map of Barley. Genetics 2000:156 1997-2005. Saghai Maroof, M. A., R. M. Biyashev, G. P. Yang, Q. Zhang and R. W. Allard, 1994 Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations and population dynamics. Proc. Natl. Acad. Sci. USA 91: 5466–5470. Thiel, T, W. Michalek, R.K. Varshney, and A. Graner. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley ( Hordeum vulgare L.). Theor. Appl. Genet. 106: 411–422.