Mathematical Modeling of Recombination Repair Mechanism
advertisement

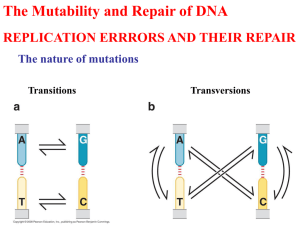
Mathematical modeling of recombination repair mechanism for double strand DNA breaks in Escherichia coli bacterial cells Alaa Mohamed Researcher Assistant, Nano Science and Technology Center, Nile University, Cairo, Egypt. Under the supervision of: Dr. Oleg V. Belov Deputy Head of the Radiobiology Department, Laboratory of Radiation Biology, Joint Institute for Nuclear Research The Project of LRB Mathematical modeling of repair systems in living organisms Dr. Oleg Belov, Laboratory of Radiation Biology UV irradiation and Mutagenesis The Project’s aim Ionizing radiation Induced DNA damages by ionizing radiation DNA Damages • • • • • Single-strand breaks Base damages Sugar damages Double-strand breaks Clustered DNA damages DNA Repair • • • • • Base excision repair Nucleotide excision repair Mismatch repair Recombination repair SOS repair The Project We quantitatively modeled the recombination repair mechanism for DNA double strands breaks, induced by ionization radiation, in Escherichia coli bacterial cells Steps for building up the model Experimental data Reaction’s code Sequence of Reactions Output Run Results 1. Sequence of Reactions 2. Setting up reaction codes dsDNA + RecBCD Un-winded DNA * RecBCD complex Un-winded DNA * RecBCD complex Single Strand DNA tail Single Strand DNA tail + RecA Activated RecA enzyme Activated RecA enzyme Single Strand DNA tail + RecA Activated RecA enzyme + H DNA D-loop D Loop Holiday Junction Holiday Junction Repaired DNA 3. Parameters / Variables • • • • dsDNA RecBCD ssDNA tail Complex • • • • RecA D-Loop Holiday Junction Repaired DNA 4. Output • All reactions were simulated using Mathematica software, using two approaches: 1. Stochastic approach 2. Deterministic approach • Outputs we obtained, characterized DNA repair steps as well as enzymes’ concentration changes. Results 1. RecBCD complex concentration changes N N t t, s t t, s Results 2. RecA enzyme concentration сhanges N N t, s t, s Results 3. D-Loop structure formation kinetics N N t, s t, s Results 4. Holiday Junction structure formation changes N N t, s t, s Results 5. Holiday Junction Vs. D-Loop N N t, s t, s Results 6. Repaired DNA formation kinetics N N t, s t, s Results 7. Repaired DNA Vs. DSB resolve N N t, s t, s Conclusion • Determined the key processes making the main contribution to the functioning of DNA recombination repair system • Developed a phenomenological model for DNA recombination repair system • Constructed a mathematical model for the DNA recombination repair system using the deterministic and stochastic approaches • Obtained and analyzed solutions for the proposed model Future tasks • Development of mathematical models for other DNA repair systems. • Development of mutagenesis model for damages induced by ionization radiations in Escherichia coli bacteria. Acknowledgments • Dr. Oleg Belov, LRB, JINR Thank You for Your Attention