DNA Mutation and Repair
advertisement

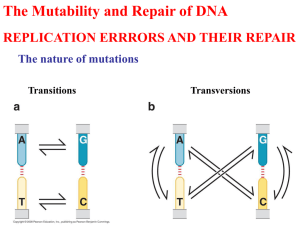
DNA Mutation and Repair Lecture 5 of Introduction to Molecular Biology 生理所 蔡少正 DNA Damage Mechanisms for maintaining genetic stability associated with DNA replication in E. Coli Machanism Cumulative error frequency Base pairing ~10-1 - 10-2 DNA polymerase actions (including base ~10-5 - 10-6 selection, 3'->5' proofreading) Accessory proteins (e.g. SSBP) ~10-7 Post-replication mismatch correction ~10-10 Spontaneous alterations: (a) Mismatches: Occurs during DNA synthesis (i.e. replication, repair, or recombination) (b) Tautomeric shifts Nucleotides spontaneously under go a transient rearrangement of bonding, e.g. a shift from NH2 (amino form) to NH (imino form) or C=O (keto) to C-OH (enol). Therefore, if any base in a template strand exists in its rare tautomeric form during DNA replication, misincorporation in the daughter strand can result. Base Pairing of Imino A-C (c) Deamination Three of the four bases normally present in DNA (cytosine, adenine, and guanine) contain amino group (NH2). The loss of the amino group (deamination) can occur spontaneously and result in the conversion of the affected bases to uracil, hypoxanthine, and xanthine, respectively. (d) Loss of bases Depurination and depyrimidination: The loss of purines or pyrimidines from DNA usually occurs at acidic pH; however, it can also happen in physiological pH (~10,000 purine per day in mammalian cell; ~500 pyrimidine/day). This will results in breaking the 3' phosphodiester bond called b-elimination. Induced Mutations (a) Physical agents that damage DNA: --- Ionizing radiation: OH, O2-, H2O2, damage base and sugar residues. --- UV radiation: Cyclobutane pyrimidine dimers, Thymidine dimers (T-T) dimer Chemical Agents (b) Chemical agents that damage DNA: --- Alkylating agents: Alkylating agents are electrophilic compounds with affinity for nucleophilic centers in organic macromolecules. These include a wide variety of chemicals, many of which are proven or suspected carcinogens (such as nitrous acid, hydroxylamine, and ethylmethane sulfonate, EMS), Adding alkyl group to hydrogenbonding oxygen of G or T, resulting in G-T mispairing G-C ---> G*T --->A-T T-A --->T*-G ---> CG Base-analogue Agents A base analogue is a substance other than a standard nucleic acid base that can be incorporated into a DNA molecule by the normal process of polymerization. Such a substance must be able to pair with the base on the complementary strand being copies, or the 3'->5' editing function will remove it. For example, 5-bromouracil is an analogue of thymine and might cause an A-T to G-C transition mutation. Base Analogue Intercalating Agents: Intercalating agents: Substances whose dimensions are roughly the same as those of a purine-pyrimidine pair. In aqueous solutions, these substances form stacked arrays, and are also able to stack with a base-pair by insertion between two base-pairs. This may result in frameshift mutation. Model of intercalating agent induced mutagenesis Metabolite Mutagens Chemicals that are metabolized to electrophilic reagents: Aflatoxins, benzo[a]pyrene A mutagen is a physical or chemical agent that causes mutations to occurs. Mutagenesis is the process of producing a mutation. Mutant refers to an organism or a gene that is different from the normal or wild type. Reversion and the Ames test: Mutants may have second mutation and become wild type again. Reversion was used as a means of detecting mutagens and carcinogensthe Ames test DNA Repair Mechanisms (1) Repair by direct reversal: The simplest mechanism. e.g. UV induced T-T dimer is recognized by photolyase and is cleaved into intact thymine (light dependent). This is called photoactivation Excision Repair (2) Excision Repair: The most ubiquitous repair mechanism, which can deal with a large variety of structural defects in DNA. Recombinational Repair (3) Recombinational repair (Postreplicational repair): Occurs before excision repair has happened or when excision repair can not fix the problem The SOS response (4) The SOS response: The SOS response system is only active in response to some signal such as a blocked of replication fork. In E. Coli, recA and lexA govern the expression of a number of other genes involved in DNA repair. This is an errorprone DNA repair mechanism and result in higher than normal mutagenesis. SOS DNA Repair 1. DNA damage 2. RecA converted to RecA* 3. RecA* facilitated LexA self-cleavage 4. Increased synthesis of SOS proteins 5. Error prone repair induced 6. DNA damage repaired 7. RecA* returned to RecA 8. LexA no longer self-cleaved 9. LexA repressed SOS genes 10. LexA repress lexA gene expression Type of Mutations(I) I. Point mutation: A. Base substitution Change in DNA Transition: One purine replaced by a different purine;or one pyrimidine replaced by a diferent pyrimidine A G T C Transversion: A purine replaced by a pyrimidine or vice versa A T C G Type of Mutations (II) Change in protein 1. Silent mutation: altered codon codes for the same a.a. GAG (Glu) --->GAA (Glu) 2. Neutral mutation: altered codon codes for functional similar a.a. GAG--->GAC (Asp) 3. Missense mutation: altered codon codes for different dissimilar a.a. GAG ---> AAG (Lys) 4. Nonsense mutation: altered codon becsomes a stop codon GAG ---> UAG (stop) Type of Mutations (III) B. Frameshift mutation: addition or deletion of one base-pair result in a shift of reading frame and alter amino acid sequence 1. Wild type: ATG ACC AGG TC Met 2. Base addition: Arg ATG ACA CAG GTC Met 3. Base deletion: Thr Thr Gln ATG ACA GGT C Met Thr Gly Val Type of Mutations (IV) II. Insertion III. Deletion IV. Translocation V. Inversion