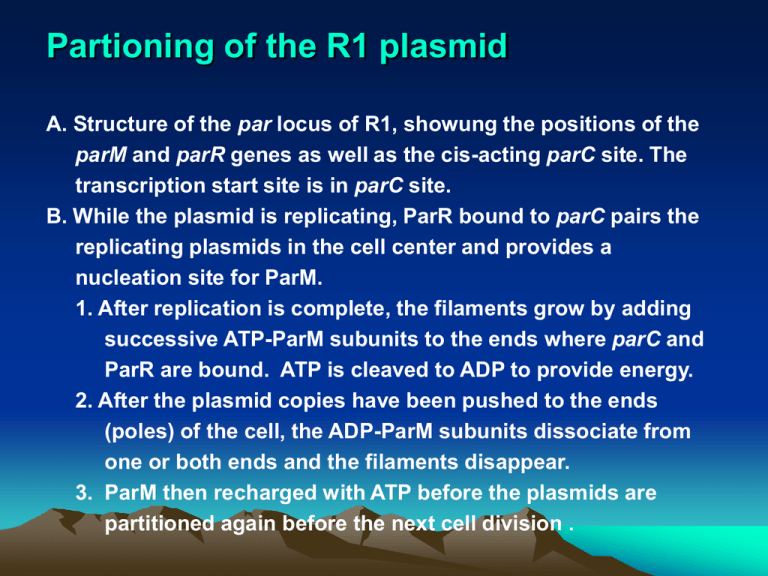
Partioning of the R1 plasmid
A. Structure of the par locus of R1, showung the positions of the
parM and parR genes as well as the cis-acting parC site. The
transcription start site is in parC site.
B. While the plasmid is replicating, ParR bound to parC pairs the
replicating plasmids in the cell center and provides a
nucleation site for ParM.
1. After replication is complete, the filaments grow by adding
successive ATP-ParM subunits to the ends where parC and
ParR are bound. ATP is cleaved to ADP to provide energy.
2. After the plasmid copies have been pushed to the ends
(poles) of the cell, the ADP-ParM subunits dissociate from
one or both ends and the filaments disappear.
3. ParM then recharged with ATP before the plasmids are
partitioned again before the next cell division .
Random curing of plasmid with no Par
system
Each plasmid has an equal chance of going to one of the
daughter cells when the cell divides, and occasional cell
inherits no plasmids.
II. Conjugation
1. Many plasmids have the ability to transfer themselves and other
DNA elements from one cell to another in a process called
conjugation.
2. Donor or male strain – the bacterial strain contains the plasmid.
In gram-negative bacteria, such cells produce a structure, called
the sex pilus, which facilitates conjugation.
3. Recipient or female strain - the bacterial strain that receives the
plasmid from a donor strain during conjugation process.
4. Many naturally occurring plasmids can transfer themselves,
called self-transmissible plasmids. Self-transmissible plasmids
encode all the functions they need to move among cells, and
sometimes they can also aid in the transfer of mobilizable
plasmids which encode some but not all of the proteins required
for transfer.
5. A plasmid will sometimes transfer into a cell that already has a
plasmid of the same Inc group, and one of the two plasmids will
subsequently be lost.
6. Mechanism of DNA transfer during conjugation in
Gram-negative bacteria
(1) F+ X F- → F+ + F+
Hfr X F- → Hfr + recombinant
(2) Conjugation is a complicated process that requires the products
of many genes: (Fig. 5.3)
i. tra genes – products required for transfer. In F plasmid, tra
genes occupy large region of the plasmid, more than 20 genes.
They can be divided into two components:
(i) Dtr component – for transfer and replication. They cluster
around the oriT.
ii. Mpf – for mating-pair formation. They consist of large
membrane-associated structure including pilus.
(i) Pilus is a tube-like structure that sticks out of the cell
surface and constructed of many copies of a single protein
called pillin.
(ii) The structure of pilus differs markedly among the plasmid
transfer system and determine the efficiency of the transfer
under various conditions. Ex., long, flexible pilus of F
A simple view of conjugation by a selftransmissible plasmid, the F plasmid
6. Mechanism of DNA transfer during
conjugation in Gram-negative bacteria
(iii) The channel – Mpf system also encodes a channel
or pore through which DNA passes during
conjugation. Little is known of its exact structure.
(iv) Coupling proteins – The Mpf component is the first
to make contact with a recipient cell. Coupling
proteins (TraD), part of the Mpf system, provide
the specificity for the transport process, so only
some of plasmid can be transferred.
(v) The relaxosome, consisting of TraY, TraM, TraI and
host-encoded IHF bound the the nicked DNA in
oriT.
(vi) The 5’ end of the nicked strand is bound to a
tyrosin in TraI and the 3’ end is associated with
TraI in an unspecified way.
6. Mechanism of DNA transfer during
conjugation in Gram-negative bacteria
iii. The central part of the Dtr component of plasmids is the
relaxase.
(i) The relaxase protein, part of relaxosome nicks DNA at nic
site in oriT sequence, is transferred into the recipient cell
along with the DNA and reseals the DNA into
single-stranded DNA.
(ii) Other proteins of relaxosome are not transferred to the
recipient cell.
(iii) Primase- Primase should not be necessary to prime the
replication in donor cell, since the free 3’ hydroxyl end of
nicked DNA can be used as primer. But the primase is
made in the donor cell, not in the recipient cell.
(iv) Why would a plasmid bother to make its own primase and
transfer it into the recipient cell if it can use the host cell
primase instead? The answer may be that it does this to
make itself more promiscuous and able to transfer into a
wider variety of bacterial species.
6. Mechanism of DNA transfer during
conjugation in Gram-negative bacteria
iv. Many naturally occurring plasmids transfer with a
high efficiency for only a short time after they are
introduced into cells and then transfer only
sporadically thereafter. Most of the time the tra
genes are repressed, and without the synthesis of
pillin and other tra gene function. Plasmids may
normally repress their tra genes to prevent infection
by male-specific phages or the cells contain plasmid
will be killed.
Partial genetic and physical map of 100
kbp F plasmid
IS2 and IS3, the insertion sequences; γδ , a transposon (Tn1000);
oriV, the origin of replication; oriT, origin of conjugal transfer; tra, the
regin encoding numerous tra functions.
The representation of the F transfer apparatus
draw from available information
The representation of the F transfer apparatus
draw from available information
1. The pilus is assembled with five TraA (pillin) subunits per turn that are
inserted into the inner membrane via TraQ and acetylated by TraX.
2. The pilus is extended through a pore constructed of TraB and TraK, a
secretion-like protein anchored to the outer membrane by the
lipoprotein TraV.
3. TraL seeds the site of pilus assembly and attracts TraC to the pilus
base, where it acts to drive assembly in an energy-dependent manner.
4. The two-way arrow indicates the opposing processes of pilus
assembly and retraction.
5. The mating-pair formation (Mpf) proteins, include G and N , which aid in
mating-pair stabilization (Mps), and S and T, which disrupt mating-pair
formation through entry and surface exclusion, respectively.
6. TraF, H, U, W and TrbC, which together with TraN are specific to F-like
systems, appear to play a role in pilus retraction, pore formation and
mating-pair stabilization.
The representation of the F transfer apparatus
draw from available information
7. The relaxosome, consisting of Y, M, l and hostencoded lHF bound to the nicked DNA in oriT is
shown interacting with the coupling protein, D,
which in turn interacts with B.
8. The 5’ end of the nicked strand is shown bound to a
tyrosine (Y) in l, and the 3’ end is shown being
associated with l in an unspecified way.
9. The retained, unnicked strand is not shown.
10. TraC, D and l (two sites for both relaxase and
helicase) have ATP utilization motifs, represented
by curved arrows.
Mechanism of
DNA transfer
during
conjugation,
showing the
mating-pair
formation (Mpf
functions) in
purple
Primase may be encoded either by the host or by the plasmid and
injected with the DNA, primes replication of the complementary strand to
make the double-stranded circular plasmid DNA in recipient.
Reactions performed by the relaxase
Fertility inhibition of the F plasmid
(regulation of the tra genes)
A. Genetic organization of the tra region.
B. The traJ product is a transcriptional activator that is required for transcription
of traY-X and finO from promoter PtraY.
C. Translation of the traJ mRNA is blocked by hybridization of an antisense RNA,
FinP, which is transcribed in the same region from the complementary strand.
Protein FinO stabilizes the FinP RNA.
Self-transmissible vs mobilizable plasmids
1. Plasmids that can not transfer themselves but can be
transferred by other plasmids are said to be moblizable,
and the process by which they are transferred is called
mobilization.
2. The simplest mobilizable plasmids merely contain the oriT
sequence of a self-transmissible plasmid can be mobilized by
that plasmid.
3. The tra genes of the Dtr system of the mobilizable plasmids
are called the mob genes and the region required for
mobilization is called the mob region.
4. Naturally occurring mobilizable plasmids can often be
mobilized by a number of tra systems.
5. Not all mobilizable plasmids can be mobilized by all selftransmissible plasmids, due to the comunnication between
the coupling protein of self-transmissible plasmids and
relaxase of mobilizable plasmids .
Integration of the F plasmid by
homologous recombination between IS2
elements in the plasmid and in the
chromosome, forming an Hfr cell
Creation of a prime factor by homologous
recombination
Prime factor –
a plasmid with
an incorporated
chromosomal
gene(s).
Selecting prime factors on the basis of
early transfer of a marker
A. The hatched region indicates F DNA.
B. A prime factor transfers the pro marker early.
Genetic mapping by Hfr crosses
• Chromosomal DNA
can be transferred
by an integrated
plasmid.
• Once in the
recipient cell, the
transferred
chromosomal DNA
may recombine with
homologous
sequences in the
recipient
chromosome.
Genetic mapping by Hfr crosses
If the trp region is
transferred from the
donor to the recipient,
it can recombine to
replace the
homologous region in
the recipient, giving
rise to a Trp+ Arg+
recombinant.
Genetic mapping by Hfr crosses
The chromosome is
transferred from the
donor to the recipient,
starting at the position
of the integrated
self-transmissible
plasmid (arrowhead).
The plating media
used to select the
markers are also
shown.
Genetic mapping by Hfr crosses
1. Large arrows indicate
the known markers
used for the Hfr gradient
of transfer.
2. The small arrows
indicate the position of
integration of the
plasmid in some Hfr
strains, including PK191
(located near the
position of hisG at 44
min).
III. Constructing a plasmid cloning
vector
1. A cloning vector is an autonomously replicating DNA into
which other DNAs can be inserted so that many copies
of original piece of DNA can be obtained.
2. Most plasmids, as they are isolated from nature, are too
large to be convenient as cloning vectors and/or often
do not contain easily selectable genes that can be used
to move them from one host to another.
3. A cloning vector can be constructed using the
techniques of genetic engineering. A cloning vector
should contain three elements: origin of replication (ori),
selection marker and unique (multiple) cloning site(s).
4. pBluescript SK as an example.
質體載體(plasmid vector)
質體載體為一個經過修飾過後的質體,將細菌自然存
在的質體移除或加入一些DNA片段而形成。
選殖載體至少必須具備三種不同功能的DNA片段:
(1) 複製的起始點(origin of DNA replication):
為質體開始複製的地方。
(2) 多選殖位點(multiple cloning sites,MCS):
為一小段DNA,內含有多個不同的限制酶的切
位,以供外源DNA片段插入。
(3) 篩選標誌(selection marker)::以 pBluescript SK
載體例,其篩選標誌為合成分解氨比黴素(ampicilin)
的基因。當大腸桿菌含有此載體時,可以在含有氨比黴
素的環境下生長。lacZ’基因為另一種常用的篩選
標誌,可用來確定是否有一段DNA插入載體之MCS內。
The rationale behind insertional inactivation of
the lacZ’ gene carried by pUC8
(a) The bacterial and
plasmid genes
complement
each other to
produce a
functional
β–galactosidase
molecule.
(b) Recombinants are
screened by plating
onto agar
containing X-gal
and IPTG.
Cloning vector pBluescript SK
Plasmid amplification
The copy number of some plasmid vectors (for
example, the pUC8 or pBluescript SK ) can be
increased by following procedures:
1. Grow the plasmid-containing bacteria to early
log phase.
2. Adding the antibiotic chlorophenicol (CM)
3. Culturing the bacteria over night.
Why?