Plasmids and Bacteriophages
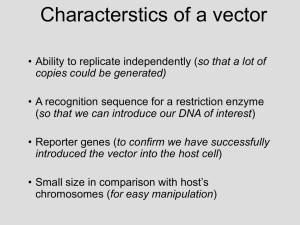
Plasmids: autonomously replicating
extrachromosomal DNA molecules present
mostly in the bacterial cells.
Bacteriophages (phages): bacterial viruses.
Plasmids
General properties of plasmids:
dsDNA, mostly circular
Size: 3 kb-150 kb
Copy number: low, intermediate, or high
Host range: narrow v.s. broad
Stability
Incompatibility
Transfer: self-transmissible;
mobilizable, nonconjugative;
nonconjugative, nonmobilizable
Features of selected plasmids of E. coli
Plasmid Size
(kb)
Copy
Conjugative
number
Other
phenotype
ColE1
6.6
10–20
No
Colicin production
and immunity
F
95
1–2
Yes
E. coli sex factor
R100
89
1–2
Yes
Antibiotic-resistance
genes
P1
90
1–2
No
Plasmid form is
prophage; produces
viral particles
R6K
40
10–20
Yes
Antibiotic-resistance
genes
Plasmid replication:
Mode of replication
Cairns intermediate (θ or butterfly form)
Rolling circle
Requirements for host enzymes
Control of copy number
Negative control
Inhibitor-target mechanism
Iteron-binding mechanism
Replication
of ColE1
Rop
Evolution of pUC18/19
ColE1
oriV
pBR322
oriT
pUC18/19
rop
Amplification
For ColE1-related plasmids, like pBR322
By chloramphenicol treatment
Copy number increases up to 1,000X
Mechanisms
Replication of host chromosomal DNA
decreases, while the machinery for plasmid
replication is more stable.
Rop concentration decreases, and that way
blocks the negative control of plasmid copy
number.
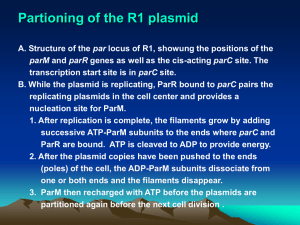
Plasmid R1
control circuit
for replication
Iteron-binding mechanism
Stability of plasmids
Partitioning
Lethal segregation
Resolution of plasmid multimers
(e.g., Xer and cer of ColE1)
Regions involved in partitioning of plasmids
Centromere-like function
The binding of ParA and ParB to parS serves as a unit for
interaction with the host components involved in segregation.
parB-mediated postsegregational killing
of plasmid free cells
of R1 plasmid
Targets of poisons: cell
membrane, gyrase,
DnaB, unknown
Antitoxins:
mRNA (type I)
Protein (type II)
h: hok (host killing) mRNA; s: sok (suppression of killing) mRNA.
Incompatibility
Incompatibility groups:
IncA-IncZ
Factors determining host range
Stable mating pair formation and
mobilization
Restriction enzymes in the recipient
Replication defect
F plasmid
Cell contact via
the sex pilus
DNA mobilization
and transfer
Surface
exclusion:
The F+ strains
do not usually
serve as the
recipient
DNA replication
while transferring
F plasmid
Excision of F plasmid from
host chromosome
Integration of F plasmid into
host chromosome
Hfr strain
Integration of
F’ plasmid
Aberrant excision of F plasmid
from host chromosome
F’ plasmid
E. coli Hfr
strains
Transfer of
chromosomal
DNA from
Hfr into a Frecipient
Transconjugant
s
Hfr x F-
Time of entry
Hfr: str-s; a+, b+, c+,
d+, e+
F- : str-r; a-, b-, c-, d-,
e-
Construction of the genetic map of E. coli by conjugation
Phages
Bacterial viruses;
replicate only within a metabolizing bacterial cell.
Structures
Coat
Nucleic acid (dsor ss-DNA or RNA;
linear or circular)
Life cycle
of phages
Lytic (virulent) phages
Lysogenic (temperate)
phages
Properties of a phage-infected bacterial culture
Multiplicity of infection (moi)
Poisson’s law
P(n)=mne-m/n!
m: moi;
n: no. absorbed phage
Plaque
Infective center
Burst size
One step growth curve of phage
Factors contributing to
host specificity:
Receptors
Ability of bacterial RNA pol to
recognize phage promoters
Host restriction and
modification
Min. after infection
Properties of several phage types
Phage
Host
DNA;
RNA
Form
fX174
E
DNA
ss, circ
-
Lysis
M13, fd, f1
E
DNA
ss, circ
-
Extrusion
Mu
E
DNA
ds, lin
+
Lysis
T7
E
DNA
ds, lin
+
Lysis
l
E
DNA
ds, lin
+
Lysis
P1
E
DNA
ds, lin
+
Lysis
SPO1
B
DNA
ds, lin
+
Lysis
T2, T4, T6
E
DNA
ds, lin
+
Lysis
MS2, Qb, f2
E
RNA
ss, lin
-
Lysis
VC
DNA
ss, cir
+
Extrusion
CTXf
Lysoginize
Mode of
release
Lytic cycle
Regulatory
cascade in
lytic cycle
Temporal control of SPO1
transcription
Control of phage l life cycle
Lytic cycle
Lysogenic cycle
Early gene
expression
Expression of late genes
Expression of repressor
Genetic map of phage l
PL
PR
PR’
cos
Temporal control of
transcription during
lytic infection by
phage l
tR1
tL1
Without N
tR2
With N
A
tR3
A
Gene expression
regulation by
antitermination
(nutL)
Antitermination by N protein
N protein is an RNA-binding protein (via an Arg-rich domain),
recognizing a stem loop formed at the nut sites.
Host proteins are involved in antitermination.
N causes antitermination at both r-dependent and r-independent
terminators by restricting the pause time at the terminator.
Antitermination by Q protein
Q binds to the qut site, which overlaps PR’, alters the RNA
pol in a way that it resumes transcription and ignores the
terminator, continuing on into the late genes.
Late gene
expression
DNA replication and maturation
Modes of replication:
q and rolling circle
Cutting and packaging of DNA (38-51 kb)
terminase (Ter system)
two cos sites
Lysogenic cycle
General properties
Turbid plagues
Immunity
Induction
Mechanism of immunity
CI repressor; OR and OL
Homoimmune and
heteroimmune
lcI -; lvir
Prophage integration
att sites (attP and attB)
Integrase (Int)
IHF (host factor)
Prophage exision
att sites (attL and attR)
Integrase
Exisionase (Xis)
IHF
Site-specific
recombination
gal
bio
Synthesis of cI:
promoters PRE and PRM
PRE
CII acts on PRE, PI and Panti-Q
CIII protects CII
from degradation
by HflA
cyL and cyR mutations prevent
establishment of lysogeny in cis
OL and OR contains three repressor-binding sites
Blocks access of RNA
pol to the promoter
AT-rich spacers allow DNA twist more
readily which enhances the affinity of
the operator for repressor
The lytic cascade
requires Cro (the
repressor for lytic
infection)
Fate of a l infection: lysis or lysogeny
CI:
OR1 > OR2 > OR3
OL1 > OL2 > OL3
Cro:
OR3 > OR2 = OR1,
OL3 > OL2 > OL1
The key to the
fate is CII
CII is degraded
by host
protease, and
stabilized by CIII
Induction of
l prophage
Transduction
Specialized transduction (e.g. l transduction)
Generalized transduction (e.g. P1 transduction)
Methods to determine plasmid copy number
Quantification of gene
products
→Activity of enzymes
→Fluorescence (GFP)
→Rate of segregation
Without separation/isolation of
plasmid DNA
→Dot blot
→Sequence-specific assay/ILA
→PCR
Quantification of nucleic acids
With separation/isolation of
plasmid DNA
→HPLC
→Density gradient centrifugation
→AGE and densitometry
→AGE and image analyzer
→AGE and Southern blot
→PCR and TGGE
→CGE
→FIA/FIP
Friehs K. 2004. Adv Biochem Engin/Biotechnol. 86: 47-82
Methods to avoid segregational plasmid instability
Adding an antibiotic to the medium
Complementation of chromosomal mutations
Post segregational killing of plasmid free cells
Influences of plasmid size and form
Active plasmid partitioning
High copy number and plasmid distribution
Lowering the difference in specific growth rates by the internal
factors
Influences of cultivation conditions (pH, O2, phosphate, etc.)
Integration into the chromosome
Friehs K. 2004. Adv Biochem Engin/Biotechnol. 86: 47-82