Chapter 11. RNA synthesis and processing (P169, sP841)
advertisement

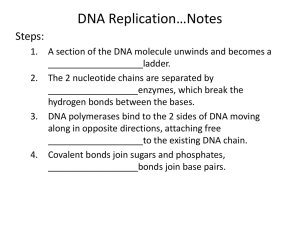
" The Central Dogma of molecular biology" replication transcription Reverse transcription translation Chapter 10 Transcription (RNA Biosynthesis) Transcription*: RNA biosynthesis from a DNA template is called transcription. transcription DNA •products:mRNA tRNA rRNA RNA Enzymes and Proteins involved in transcription : •substrates : NTP ( GTP, CTP ATP, ) •template: DNA • enzyme : RNA polymerase • the other Protein factors UTP, Chemical reaction-- polymerization reaction: RNA polymerase catalyze formation of Phosphodiester bonds and release pyrophosphate (ppi) RNA polymerase RNA precursor RNA biosynthesis is similar to DNA biosynthesis*: Template- DNA Enzyme—dependent on DNA Chemical reaction--the formation of Phosphodiester bonds Direction of synthesis--- 5’ obey the ruler of base paired 3’ RNA biosynthesis includes three stages: Initiation: RNA polymerase binds to the promoter of DNA, and then a transcription “bubble” is formed. Elongation: the polymerase catalyzes formation of 3’5’-phosphodiester bonds in 5’3’ direction, using NTP as building units. Termination: when the polymerase reaches a termination sequence on DNA, the reaction stops and the newly synthesized RNA is released. Formation of a transcription bubble 1. RNA biosynthesis in prokaryotes RNA polymerase in E. coli : consists of five subunits, a2bb’ws, which is called “holoenzyme”. The s subunit functions as a starting factor that can recognize and bind to the promoter site. The rest of the enzyme, a2bb’w, is known as “core enzyme”, responsible for elongation of the RNA sequence. E. coli RNA polymerase b b b b a a Core enzyme a s a a Holoenzyme 1) Important terms in RNA biosynthesis. A)Operon*: a coordinated unit of gene expression, which usually contains a regulator gene and a set of structural genes. B) Promoter site*: a region of DNA templates that specifically binds RNA polymerase and determines where transcription begins. regulator gene 5 3 Promoter site structural genes RNA-pol 3 5 The –10 sequence: refers to the consensus TATAAT, and is known as “Pribnow box”. The –35 sequence: refers to the consensus TTGACA, which is recognized by the s subunit of RNA polymerase, DNA template -35 TTGACA -10 TATAAT Pribnow box recognition site +1 RNA 5 3 -5 0 -40 -30 -20 -10 1 10 3 5 consensus -35 sequences region the site of transcription (the start site) -10 region TTGACA AA C T G T recognition site T A T A A T Pu A T A T T A Py (Pribnow box) C) Sense and antisense strand: The antisense (-) strand refers to the DNA strand that is used as template to synthesize mRNA. The sense (+) strand of a DNA double helix is the non-template strand that has the same sequence as that of the RNA transcript except for T in place of U. Antisense (-) strand = template strand Sense (+) strand = coding strand Sense and antisense strand: coding strand antisense strand 5 3 3 5 template strand sense strand 3) Process of RNA biosynthesis: The process is similar to DNA synthesis but no primer is needed and T is replaced by U. A) Initiation: •σ factor recognizes the initiation site(-35 region), the holoenzyme of RNA-pol bind to duplex DNA and move along the double helix towards –10 region. •the holoenzyme of RNA-pol arrived on –10 region,and bind to –10 region ,DNA is partially unwound and was opened 10-20 bp length. •Then incoming 2 neighbour nucleotides which base pairs are complementary with DNA template, RNA polymerase catalyzed the first polymerization reaction. 5’-pppG -OH + NTP – 5’ -pppGpN – OH + ppi s pppG NTP- OH pppGpN ppi initiation complex: RNApol(α2ββˊσ)-DNA-pppGpN-OH3’ DNA template TTGACA TATAAT DNA template TTGACA TATAAT DNA template TTGACA TATAAT DNA template TTGACA TATAAT + “Core” s The first phosphodiester bond formed B) Elongation: after the first phosphodiester bond has been formed, the s subunit is released. The core enzyme moves in a 5’3’ direction on the DNA strand while it is catalyzing elongation of the RNA transcript. DNA template TTGACA TATAAT DNA template TTGACA TATAAT NTP DNA template TTGACA TATAAT tanscription complex: RNA-pol (core enzyme) ···· DNA ···· RNA RNA polymerase Sense strand Direction of transcription Rewinding 3’ 5’ 5’ 3’ 5’PPP Unwinding Antisense strand Newly synthesized RNA strand C) Termination: when the core enzyme reaches a termination sequence, the region near the 3’end of RNA forms a hairpin structure by self base-pairing. The transcription stops, the core enzyme and the newly synthesized RNA are released. For those DNA templates that lack the sequence to produce a hairpin structure of the RNA transcript, a protein factor called “r ” recognizes the termination site, stops transcription, and causes release of the newly synthesized RNA. A hairpin structure at the 3’end of RNA C U U G G G•C A•U C•G C•G G•C C•G C•G G•C 5’ A-U-U-U-U-OH 3’ Termination by hairpin structure of RNA RNA-polymerase 5 3 5’pppG 3 5 DNA 5 RNA 3 RNA polymerase Ribosome The multiple-site transcription in bacteria Subunits of RNA polymerase in E. coli Subunit Size (AA) Function a 329 required for assembly of the enzyme; interacts with some regulatory proteins; involved in catalysis b 1342 involved in catalysis: chain initiation and elongation b' 1407 binds to the DNA template s 613 directs the enzyme to the promoter w 91 required to restore denatured RNA polymerase in vitro to its fully functional form 4) Post-transcriptional modification: The newly synthesized precursors of rRNA and tRNA in bacteria undergo a series of process. A) Processing of rRNA: the 16S, 23S, and 5S rRNAs in prokaryotes are produced by cleavage of a rRNA precursor, catalyzed by ribonuclease III. Additional processes include methylation of bases and sugar moieties of some nucleotides. Processing of rRNAs 16S 23S 5S First cleavage 16S 23S 5S Second cleavage 16S(1.5kb) 23S(2.9kb) 5S(0.12kb) B) Processing of tRNA: The removal of the 5’ end of tRNA precursors is catalyzed by RNase P. RNase P is a ribozyme consisting of RNA that possesses enzyme activity. Other processes include the addition of nucleotides (CCA) to the 3’-end of tRNA, and formation of some unusual residues such as pseudo-U, I, T, methyl-G, and DHU, etc. Modification of some residues in tRNAs 4) Inhibition of transcription: Rifampicin: an antibiotic that specifically inhibits the initiation of transcription by blocking the formation of the first several phosphodiester bonds in RNA biosynthesis. Streptolydigin: binds to bacterial RNA polymerase and inhibits elongation of RNA chain. Actinomycin D: binds to DNA and prevents transcription (at low concentrations it doesn't affect DNA replication) 2. RNA biosynthesis in eukaryotes 1) RNA polymerases in eukaryotes: three enzymes, each of which contains 12 or more subunits. Polymerase Pol I Pol II Pol III location nucleolus nucleoplasm nucleoplasm RNAs transcribed 28S, 18S, 5.8S rRNA pre-mRNA, snRNA tRNA, 5S rRNA, U6 snRNA, 7S RNA 2) Process of eukaryotic RNA synthesis A) Initiation: similar to Pribnow box, a start site consensus (called TATA box) at –25 is required for the recognition by RNA polymerase in eukaryotes. A A TATA A T T -25 Structural gene +1 Pol II requires several transcription factors to start transcription: TFII-A: to stabilize the TFIID-TATA box complex; TFII-B: to link Pol II to the initiation complex; TFII-D: to recognize and bind to the TATA box; TFII-E: to interact with Pol II and TFII-B; TFII-F: to form Pol II-TFIIF complex. It also has DNA helicase activity; TFII-H, -J: to form the initiation complex. TATA box Structural gene S TFII-D TBP S TBP A B S A TBP B Pol II F F S A TBP B E A S TBP FE H B J H J B) Elongation : after the initiation complex has formed, the RNA polymerase catalyzes transcription in a 5’3’direction, using the (-) DNA strand as template. Soon after the 5’end of the extending RNA chain appears from the polymerase complex, a cap structure is added at the end. Cap structure of mRNA O CH3 N+ N N HN H2N O - O P O CH2 O O - O H P O H O H O - 7-methylguanylate H OH P OH O O CH2 O H Base H H H O - O P OCH3 O O CH2 O H H Base H H O OCH3 C) Termination: Two mechanisms may cause termination of RNA transcription: A hairpin structure formed at the 3’end of the nascent RNA causes stop of transcription, as is seen in the prokaryotic RNA synthesis. A stop signal sequence, AAUAAA, near the 3’end results in the recognition and binding by a specific endonuclease, which cleaves the nascent RNA chain and stops transcription. The newly synthesized mRNA precursor is then added a poly A tail by poly A polymerase. Cleavage and polyadenylation of a mRNA precursor RNA polymerase Template DNA AAUAAA Nascent RNA Cleavage signal endonuclease ATP Poly A polymerase PPi 5’ AAUAAA mRNA precursor AAAA(A)n-OH 3’ 3) Processing of eukaryotic RNA precursors: A) Gene organization: protein-coding genes in eukaryotic DNA are organized in a discontinuous fashion. The protein-coding sections are called “exons”, which are interrupted by noncoding sections called “introns”. Exon 1 Promoter Transcription initiation site Exon 1 Intron 1 Exon 1 Intron 2 Transcription Termination region B) RNA splicing: a process in which introns of a premRNA are removed to produce a functional mRNA. Exon 1 Promoter Exon 2 Intron 1 Exon 3 Intron 2 Transcription Exon 1 Exon 2 Exon 3 5’ AA(A)250 3’ Intron 1 Intron 2 RNA splicing 5’ AA(A)250 3’ C) Steps in RNA splicing: usually the exonintron boundaries are marked by specific sequences. The intron starts with GU and ends with AG. Intron 3’ splice site 5’ splice site Exon 1 GU CURAY Branch point sequence U/C11 Polypyrimidine tract AG Exon 2 I. II. Formation of a lariat intermediate: the phosphodiester bond of the 5’ splice site is attacked by the 2’-OH of the residue A in the branch point, forming a 2’5’bond and releasing the exon 1 with a new 3’OH end. Connection of exons: The new 3’-OH end attacks the phosphodiester bond at the 3’splice site causing the two exons to join and releasing the intron. RNA splicing requires the small nuclear ribonucleoprotein particles (snRNP), each of which consists of a small nuclear RNA and several proteins. They are named U1, U2, U3…. snRNPs bind to the pre-mRNA to form a complex, called spliceosome, which brings the two neighbored exons together for splicing. Exon 1 GU A AG Exon 2 snRNPs U1 U2 U1 U2 Exon 1 GU A AG Exon 2 U4-U5-U6 U1 Exon 1 GU U5 U2 A U6 U4 AG Exon 2 Spliceosome U1 Exon 1 GU U5 U2 A U6 U4 AG Exon 2 U1 Exon 1 GU U5 U2 A U6 U4 AG Exon 2 Lariat intermediate U1 Exon 1 Exon 2 GU U5 U2 A U6 U4 AG-OH 3’ 4) Alternative processing: A) Alternative polyadenylation sites: this will cause different splice-sites and produce different mRNAs with varied lifetimes. Poly A Exon 1 Exon 2 Splicing Poly A Exon 3 B) Alternative splicing: will cause different combinations of exons from a primary transcript of a single gene. This may be resulted from regulatory proteins that control the use of certain splice-sites. Exon 1 Exon 2 Splicing Exon 3 5) RNA editing: refers to the reactions that can change the nucleotide sequence of an mRNA molecule by non-splicing mechanisms. The change may include: nucleotide(s) change, deletion, and insertion. e.g. the mRNA for apolipoprotein B in the liver is translated to apolipoprotein B100, while in the small intestine the mRNA is changed to yield a new termination codon (UAA), resulting in a much shorter protein, apolipoprotein B48. Apolipoprotein B mRNA CAA n Editing (deamination) Tr an s lat io NH4 UAA Edited mRNA translation ApoB100 ApoB48 Lipoprotein assembly LDL receptor binding Lipoprotein assembly 3. Reverse transcription and RNA replication 1) Reverse transcription: biosynthesis of DNA using RNA as a template. It is important for some viral infections. These viruses are called retroviruses, such as some tumor viruses and HIV. Reverse transcription is also a powerful tool in molecular biological techniques or genetic engineering, such as RT-PCR. 2) RNA replication: RNA replication occurs in some viruses. These viruses encode RNA-directed RNA polymerase that catalyzes biosynthesis of RNA from an RNA template. RNA replication helps the RNA viruses easily reproduce their progeny viruses.