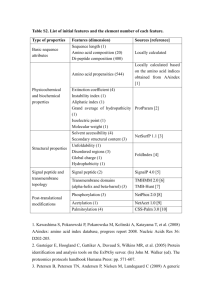
Not predictable…
advertisement

Corrections SEQUENCE 4 • • • • • • • • • • • • • • • • • • • • • • • • >seq4 MSTNNYQTLSQNKADRMGPGGSRRPRNSQHATASTPSASSCKEQQKDVEH EFDIIAYKTTFWRTFFFYALSFGTCGIFRLFLHWFPKRLIQFRGKRCSVE NADLVLVVDNHNRYDICNVYYRNKSGTDHTVVANTDGNLAELDELRWFKY RKLQYTWIDGEWSTPSRAYSHVTPENLASSAPTTGLKADDVALRRTYFGP NVMPVKLSPFYELVYKEVLSPFYIFQAISVTVWYIDDYVWYAALIIVMSL YSVIMTLRQTRSQQRRLQSMVVEHDEVQVIRENGRVLTLDSSEIVPGDVL VIPPQGCMMYCDAVLLNGTCIVNESMLTGESIPITKSAISDDGHEKIFSI DKHGKNIIFNGTKVLQTKYYKGQNVKALVIRTAYSTTKGQLIRAIMYPKP ADFKFFRELMKFIGVLAIVAFFGFMYTSFILFYRGSSIGKIIIRALDLVT IVVPPALPAVMGIGIFYAQRRLRQKSIYCISPTTINTCGAIDVVCFDKTG TLTEDGLDFYALRVVNDAKIGDNIVQIAANDSCQNVVRAIATCHTLSKIN NELHGDPLDVIMFEQTGYSLEEDDSESHESIESIQPILIRPPKDSSLPDC QIVKQFTFSSGLQRQSVIVTEEDSMKAYCKGSPEMIMSLCRPETVPENFH DIVEEYSQHGYRLIAVAEKELVVGSEVQKTPRQSIECDLTLIGLVALENR LKPVTTEVIQKLNEANIRSVMVTGDNLLTALSVARECGIIVPNKSAYLIE HENGVVDRRGRTVLTIREKEDHHTERQPKIVDLTKMTNKDCQFAISGSTF SVVTHEYPDLLDQLVLVCNVFARMAPEQKQLLVEHLQDVGQTVAMCGDGA NDCAALKAAHAGISLSEAEASIAAPFTSKVADIRCVITLISEGRAALVTS YSAFLCMAGYSLTQFISILLLYWIATSYSQMQFLFIDIAIVTNLAFLSSK TRAHKELASTPPPTSILSTASMVSLFGQLAIGGMAQVAVFCLITMQSWFI PFMPTHHDNDEDRKSLQGTAIFYVSLFHYIVLYFVFAAGPPYRASIASNK AFLISMIGVTVTCIAIVVFYVTPIQYFLGCLQMPQEFRFIILAVATVTAV ISIIYDRCVDWISERLREKIRQRRKGA Compute pI/Mw tool !!! If you choose the wrong format for the sequence… With the correct format: ProtParam • SAPS SAPS (1) SAPS (2) doi: 10.1093/bioinformatics/bti797 http://en.wikipedia.org/wiki/Coiled_coil The coiled-coil domains are annotated according to 3D structure data (experimental data) Coiled-coil prediction • Coils Coils prediction http://www.ch.embnet.org/software/COILS_form.html Coiled-coil prediction • PairCoil (not always working…) Paircoil prediction Coiled-coil prediction • PairCoil2 Parcoil2 results Coiled-coil prediction • Sliding window (Protscale) Sliding window amino acid scaleexample: • Bad results---- Bad results…. Sliding windows and amino acid scales Transmembrane domain: alpha-helix of 20 amino acids (hydrophobic) • -> amino acid scales: hydrophobicity and alpha helix • -> sliding window size: 20 amino acids Protscale Amino acid scale: Kyte and Doolittle (hydrophobicity) Sliding window size: 21 amino acids Protscale Amino acid scale: Chou&Fasman (alpha helix) Sliding window size: 21 Sliding windows and amino acid scales Transmembrane domain: alpha-helix of 20 amino acids (hydrophobic) • -> amino acid scales: hydrophobicity and alpha helix • -> sliding window size: 20 amino acids Method based HMM or NN HMMTOP HMMTOP • Protein: seq4 • Length: 1127 • N-terminus: IN • Number of transmembrane helices: 8 Transmembrane helices: 65-82 409-432 445-468 916-940 970-993 1020-1039 10521072 1089-1106 TMHMM (1) TMHMM (2) TMpred (1) PSORT II (1) - Look for the presence of a signal peptide. No signal peptide Signal peptides are often predicted as ‘transmembrane’ domains (or vice versa) as they amino acids with similar biochemical properties (hydrophic and alpha helix). Transmembrane: resume HMMTOP (8 TM) 1 in Big loop 1130 PSORT II (10 TM) 1 1130 Tmpred (10 TM) 1 in 1130 TMHMM (11 TM) 1 out 1130 ? missed TM P39986 O14072 Q9HD20 Q9Y139 P90747 Q9LT02 Q12697 O74431 Q9N323 Q27533 Q21286 Q9NQ11 O14022 Q95050 Q9XXW1 Consensus Segment Helix ...........mtkksfvsspivrdstllvpksliakpyvlpffplyatfaqlyfqqydryikgpewtfvylgtlvslnilvmLmpaW.nvkikakfny......... ...........mgskalitspdissgqlyiklptffhlyvwpfalfvypyigyvyqnklyseevryltyiavg...tihalfwLageW.ntkvyclmtc......... ..........................................................................................mekweelnshqp...... lvqyvslhvriptpltgvvlpfvplylsafylwinvtggqendttnndvitadnqtttdnittwndvgfigvlaiaflhiltlLfcyW.svhvlafltc......... ................mgvdqlvetiipynlrsiathlyvppftiitaiwtyvwlnifgyeey.yelgmlgyaaifvilalvlLfchW.mmpvrcflmc......... ............mssfrvggkvvekvdlcrkkqlvwrldvwpfailytvwlttivpsidfsd.....acialgglsafhilvlLfttW.svdfkcfvqfskmnlellv rsasennrgsfsghddvhnqhseylkpdyhekfypqyapnlhyqrfyiaeedlvigiaayqts.kfwyiiynlccfltfglvyLltrW.lphlkvkly.......... ngsgvysdeeeitemmleelnihpvlrresvgeaaglsedgccqilylveedlevgiagyktn.ksryrlyqaiclltlglayLifrW.lpkyfirfv.......... ...mstnnyqtlsqnkadrmgpggsrrprnsqhatastpsassckeqqkdvehefdiiayktt.fwrtfffyalsfgtcgifrLflhW.fpkrliqfr.......... .............................................mtlesgdhtltlfayrtg.pfrtilfyaltvltlgifrLilhW.kqkwdvkmr.......... mtsereplldtttrnrvydttdnpstkimkrekdnpkakttsfnqgklnigeetcdlyayket.igrqilfwlltivtlgfyqLlayW.vkslfvkvr.......... ....................msadssplvgstptgygtltigtsidplsssvssvrlsgycgs.pwrvigyhvvvwmmagiplLlfrW.kplwgvrlr.......... ...................mdsielkqlvpendsepgtprqllfqhydisneetigikpfksi.pakvyilrvteiltlgllhLiltW.lpefrlkwi.......... ...........................mrvssieaemenpidvdktdvegelkikqvtllren.........ivkkivfflvaifcsd.rpsvlkkvfy......... aleffiffllsltitygiliirkhiqslflkpsllkdsdyviiytineeyntfyntnyfkkyishinhmihtfikkkkknikknikkWnfqkynilflqfvcnlldii -----------------------------------------------------------------------------------L---W--------------------------------------------------------------------------------------------------------------------------------------------------------------aaaaaaaaaaaaaaaaaaaaaa-------------bbbbbbbbbbbbbbbbbbbbb-------------------- • The protein is known to contain 12 TM: one TM is missing at the N-terminus • The possible ways to find the correct protein topology is to do a multiple alignment with other family members, or to do some 3D experiment (which are difficult with proteins containing transmembrane domains) SEQ4 = Q9N323 Kristian Axelsen: personnal communication P0AER0 The Aquaglyceroporin contains ½ transmembrane regions which can not be predicted by programs, because the region is too short (less than 20 amino acids). There is no way to predict such transmembrane regions, except by doing 3D experiments. 3D experiments is the only way to confirm and ‘predict’ correctly transmembrane domains. Similarity analysis could then help to predict such regions in other protein of the same family. M3 and M7 are ‘demi’ transmembrane: not predictable http://www.uniprot.org/uniprot/P0AER0#section_features Look for the transmembrane regions of P31243 (try the different transmembrane prediction programs): your conclusions ? No transmembrane domains are found by any program because this protein, a porin, is anchored in the membrane by a specific 3D structure called beta barrel which does not have any alpha helix…. ‘beta barrel’ Mainly composed of beta-sheets in a 16-stranded beta-barrel formation and forms a pore in the membrane 1.7 - 2.5 nm in diameter. Note that the orientation of the strands is such that side chains alternately point into the interior and exterior of the pore; the former are strongly polar residues while the latter are very hydrophobic. Beta barrel Porin from Rhodobacter Alignment of the 2 isoforms The gene has two in-frame initiation codons and two different proteins are made by alternative initiation (of translation) According to this publication (PubMed: 11274159), there is a 'Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two inframe initiation codons'. Immunoblot analysis of Protox II in spinach leaf. Total leaf Watanabe N et al. J. Biol. Chem. 2001;276:20474-20481 ©2001 by American Society for Biochemistry and Molecular Biology chloro mito Q94IG7 – Long isoform • wolfPSORT: chloroplast • TargetP: chloroplast • CH score: 0.826 • MI score: 0.026 • ER score: 0.101 • Other location: 0.060 • SignalP-NN: not secreted • score (D): 0.285 • SignalP-HMM: not secreted • SP probability: 6.2% • SA probability: 0.2% • ChloroP: chloroplast • prediction score: 0.549 • MITOPROT: mitochondria !!! exported to mitochondria with a probability of 0.71 !!!! Q94IG7 – Short isoform • wolfPSORT: mitochondrial • TargetP: mitochondrial • CH score: 0.123 • MI score: 0.504 • ER score: 0.048 • Other location: 0.400 • SignalP-NN: not secreted • score (D): 0.195 • SignalP-HMM: not secreted • SP probability: 3.1% • SA probability: 5% • ChloroP: not in chloroplast • prediction score: 0.473 • MITOPROT: other location • exported to mitochondria with a probability of 0.33 !!!!!! Cystein (61 modifications) and serine (46 modifications) are the amino acids with the highest number of known associated PTM. Beware: Resid considers the selenocystein as a PTM…this is not the case ! Phosphorylation P03372 P03372 P03372 UniProt data: Experimentally proved http://www.phosphosite.org/proteinAction.do?id=968&showAllSites=true The phosphorylation sites are localized on the ‘surface’ of the protein (homodimer) (where the amino acid are accessible to the kinases !) O-glycosylation P02724 Myristoylation P51876 P51876 NMT Myristoylator (Not predictable…) Protein: secreted protein (P02751, fibronectin) (predictable…) (predictable…) (Not predictable…) Can be predicted: -Subcellular location (PSORT, TargetP) -Domains (InterPro) -Signal -Sulfation -N-glycosylation -O-glycosylation -Phosphorylation (Not predictable…) THE END