M. guttatus - Biology Department | UNC Chapel Hill
advertisement

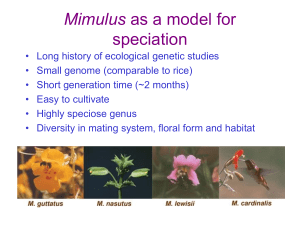
Genetics of speciation in Mimulus Todd Vision, Department of Biology, University of North Carolina at Chapel Hill, tjv@email.unc.edu Overview The plant genus Mimulus (monkeyflower) has been intensively studied by evolutionary geneticists interested in species differences for over 50 years (Vickery 1951). Recently, Quantitative Trait Locus (QTL) mapping studies have identified loci underlying reproductive isolation in two different sections of the genus. We are part of a research consortium that is working to identify the genes that underlie these QTL using the tools of comparative genomics and population genetics, and to understand the evolutionary history and ecological significance of these loci. Reproductive isolation via pollinator discrimination Mimulus lewisii and M. cardinalis are closely related and highly interfertile sister species with adjoining geographic distributions in the Sierra Nevada Mountains of California. They show strong differences in pollination syndrome and habitat preference, and hybrids are rare. M. lewisii is found at higher elevations and is almost exclusively bee-pollinated. M. cardinalis is at lower elevations and is pollinated by hummingbirds. Field studies in an area of sympatry at Yosemite on segregating F2 progeny (Figure 1) have shown that pollinators discriminate on the basis of petal size, pigmentation and nectar volume (Schemske and Bradshaw 1999). An F2 linkage map was generated using RAPD markers. QTL mapping revealed that major genes underlie the majority of flower traits that distinguish these species (Bradshaw et al. 1995, 1998). Substitution of the major anthocyanin QTL (yup) from M. cardinalis into M. lewisii reduced bee visitation by 80%. Similarly, an allelic substitution from M. lewisii into M. cardinalis at the major nectar QTL (NEC1) reduced hummingbird visitation by 50%. Thus, these two loci together account for much of the reproductive isolation between the two species. Nearly isogenic lines (NIL) are being created that “Mendelize” these loci and make them amenable to positional cloning. Fig 1.M. lewisii (A), an F1 hybrid (B), M. cardinalis (C), and examples of variation in floral traits found in F2 hybrids (D-L). From Schemske and Bradshaw (1999). Fig 3. QTL map for 7 floral traits in 526 M. guttatus x nasutus F2 progeny. Dashed line = 0.05 significance threshold. 24 minor QTL were found on the 14 linkage groups. From Fishman and Willis (2002). Reproductive isolation via hybrid sterility Species in the M. guttatus complex are exceptionally diverse in habitat and mating system. Fishman and Willis (2001) have mapped the QTL underlying phenotypic divergence and intrinsic reproductive isolation between M. guttatus (the most common species in the genus and a showy-flowered outcrosser) and M. nasutus (a selfing species with small, often cleistogamous, flowers, Figure 2) using AFLP and microsatellite markers. Differences in floral morphology are underlain by many minor QTL. However, one pair of epistatically interacting nuclear loci cause complete pollen sterility in 1/8 of the hybrid F2 offspring, consistent with the classic speciation model of Dobzhansky and Muller. An additional cytonuclear incompatibility system causes complete male sterility in ¼ of offspring carrying the M. guttatus cytoplasm. Both Dobzhansky-Muller factors and the nuclear restorer locus of the cytonuclear incompatibility are being isolated in NIL. Fig 2. M. nasutus growing in a mixed clump with M. guttatus in Shirley Creek, Calaveras Co, California. The red lines indicate M. nasutus flowers (photo: Mark MacNair) Genomic tools for positional cloning Positional cloning of the QTL in these two species pairs will be facilitated by generating genetic and physical maps that are anchored by comparative mapping markers (see next section). 30,000 Expressed Sequence Tags (EST) are being generated from M. guttatus. Alignment of these sequences against genomic DNA from Arabidopsis thaliana allows us to design PCR primers that flank introns in Mimulus. The resulting length and sequence polymorphisms are being used to score 1000 markers (using fluorescent capillary electrophoresis) in two mapping populations: 500 clonally propgated F2 progeny from M. guttatus x nasutus and 500 Recombinant Inbred Lines (RIL) from M. lewisii x cardinalis. From these data, we will obtain high-resolution linkage maps (Vision et al 2000) that can be aligned with physical maps for these species. For physical mapping, two 11X Bacterial Artificial Chromosomes (BAC) libraries have been constructed for M. guttatus (38K BAC with an average insert size of 120kb) and a similar library is being constructed for M. lewisii. These are being fingerprinted to generate a minimal tiling path, the clones of which will then be end-sequenced. The markers on the genetic map will then be localized to tiling path clones using a pooled overgo mapping strategy. Clones found to contain QTL will be shotgun sequenced in their entirety. In addition, transformation protocols are being developed to allow transgenic testing of candidate QTL. Comparative mapping The markers used to align the physical and genetic maps are homologous to known genes, and so may be used to align the maps of Mimulus and other genetic model species (particularly Arabidopsis thaliana and the more closely related Lycopersicon esculentum, or tomato), for which genome sequence is now or will soon be available. By comparison of conserved chromosomal segments among these species using the FISH software package and the Phytome database, both developed in our laboratory (Calabrese et al. 2003), the gene content of a given region of Mimulus chromosome can be predicted (Ku et al. 2000). These predictions will be used to infer candidate genes and to design markers for fine-mapping. Population genetic history of QTL regions One currently popular strategy for fine-mapping QTL is to survey the pattern of genetic variability and linkage disequilibrium along the chromosomal region of interest (Schlotterer 1999). Population genetic theory tells us that if a locus has been under directional selection, genetic variability will be reduced and linkage disequilibrium will be locally strong in the neighborhood of the causative polymorphism. From such data, we may also infer of the strength of selection, the age of the mutation, and the biogeographic history of the alleles, providing insight into “why” and “how” such speciation genes arise. To obtain these data in Mimulus, we will survey variable microsatellites obtained from BAC end-sequencing. Literature Cited Bradshaw HD, Wilbert SM, Otto KG, Schemske DW (1995) Nature 376:762-765 Bradshaw HD, Otto KG, Frewen BE, McKay JK, Schemske DW (1998) Genetics 149:367-382 Calabrese PP, Chakravarty S, Vision TJ (2003) Bioinformatics 19:i74-i80 Fishman L, Willis JH (2001) Evolution 55:1932-1942 Fishman L, Willis JH (2002) Evolution 56: 2138–2155 Ku H-M, Vision TJ, Liu J, Tanksley SD. (2000) PNAS 97:9121–9126 Schemske DW, Bradshaw HD (1999) PNAS 96: 11910-11915 Schlotterer C (2003) Trends in Genetics 19, 32-38 Vickery RK (1951) Carnegie Inst Wash. Yearb. 50:118-119 Vision TJ, Brown DG, Shmoys DB, Durrett RT, Tanksley SD (2000) Genetics155:407-420 Project Participants Clemson University Genome Institute: Fred Tompkins Duke University: John Willis, Fred Dietrich Michigan State University: Doug Schemske University of Montana: Lila Fishman University of North Carolina at Chapel Hill: Todd Vision University of Washington: Toby Bradshaw Supported by the National Science Foundation (Frontiers in Integrative Biological Research program), DBI-022731