Bayesian Variable Selection in
Semiparametric Regression
Modeling with Applications to
Genetic Mappping
Fei Zou
Department of Biostatistics
University of North Carolina-Chapel Hill
Email: fzou@bios.unc.edu
June 2012 Finland
http://www.cs.unc.edu/Courses/comp590-090-f06/Slides/CSclass_Threadgill.ppt
The Central Dogma of Molecular Biology
tall
short
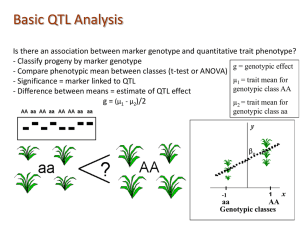
•Significant difference in genotype distributions?
http://psb.stanford.edu/psb06/presentations/association_mapping.pdf
• Copied (with modifications) from psb.stanford.edu/psb06/presentations/association_mapping.pdf
Mendel’s
Experiment
Experimental Crosses: F2
P1
Parents
P2
Experimental Crosses
•
P1
F2
AA
F1
F2:
P1
BB
P1
F1
F1
AA
AB
AB
P2
AA
BB
AB
BB
Backcross(BC)
P2
AB
BC:
AB
AB
AA
AB
F2 Data Format
0: homozygous AA, 2: homozygous BB,
1: heterozygote AB.
Data Structure
• For each subject i (i=1,2,…,n)
– Phenotype: yi
– Genotypes: xij (coded as 0, 1, 2 for genotypes
AA, AB and BB, respectively) at marker j
(j=1,2,…,m)
– Genetic map: locations of markers
– Other non-genetic covariates, such as age,
sex, environmental conditions
Locations of markers
Linkage Analysis
• Quantitative trait loci (QTL): a particular region of
the genome containing one or more genes that
are associated with the trait being assayed or
measured
QTL Mapping of Experimental
Crosses
• Single QTL Mapping
• Single marker analysis
• Interval mapping: Lander & Botstein (1989,
Genetics)
• Multiple QTL mapping
• Composite interval mapping
• Multiple interval mapping
• Bayesian analysis
Single Marker Analysis
Correlations of marker
genotypes in experimental
crosses
Interval Mapping
• Traditional QTL mapping method
• Treat QTL position as unknown and use marker
genotypes to infer conditional probabilities of QTL
genotypes
• Profile LOD scores calculated across whole genome
– LOD score is a measure for strength of support for QTL
– LOD = LRT/4.8
– In any region where the profile exceeds a (genome-wide)
significance threshold, a QTL is declared at the position with the
highest LOD score.
Profile LOD
8
lod
6
4
2
0
1
2
3 4
5
6 7 8 9 10 11 12 13 14 15 16171819 X
Chromosome
QTL
• Old believe: one trait one gene
– very unlikely
• Most traits have a significant
environmental exposure component
• The vast majority of biological traits are
caused by complex polygenic interactions
– also context dependent
Multiple QTL Mapping
• Most complicated traits are caused by
multiple (potentially interacting) genes,
which also interact with environmental
stimuli
• Single QTL interval mapping
– Ghost QTL
– Low power
if multiple QTLs affect the trait
Two QTL Data
Two QTL with opposite effects
Two QTL with effects in same direction
Multiple QTL Mapping
• Available Methods
– Composite interval mapping: searching for a putative QTL in a
given region while simultaneously fitting partial regression
coefficients for "background markers" to adjust the effects of
other QTLs outside the region
• which background markers to include; window size etc
– Multiple interval mapping: fitting multiple QTLs simultaneously
• Computationally very intensive; how many QTLs to fit?
Multiple QTL Mapping
Multiple QTL Mapping
Multiple QTL Mapping
Bayesian QTL Mapping
• Reversible jump Markov chain Monte Carlo
(MCMC) (Green 1995): treat the number of
QTLs as a parameter
– Change of dimensionality, the acceptance probability for
such dimension change, which in practice, may not be
handled correctly (Ven 2004)
• Bayesian variable selection procedures
– composite model space (Yi 2004)
– stochastic search variable selection (SSVS) (George and
McCulloch 1993)