ppt
advertisement

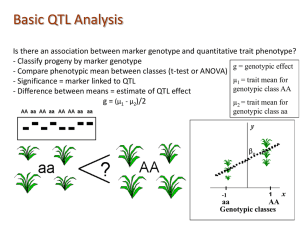
Lecture 27 : Asscociation Genetics April 21, 2014 Announcements Final exam April 29 at 3 pm, 3306 LSB (computer lab) Review session on Friday Bring questions Final lab on Wednesday Course evaluations Extra credit opportunity: earn up to 10 points for lab report Due at final exam Last Time Sequence data and quantification of variation Sequence-based tests of neutrality Ewens-Watterson Test Tajima’s D Hudson-Kreitman-Aguade Test Synonymous versus Nonsynonymous substitutions McDonald-Kreitman Today Quantitative traits Genetic basis Heritability Linking phenotype to genotype QTL analysis introduction Limitations of QTL Association genetics Mendelian trait Individual 1 2 3 4 5 6 7 8 9 10 Allele A1 Allele A2 Genotype = 12 11 22 22 11 22 12 11 22 12 Quantitative trait 16 28 40 52 Height 64 76 88 Courtesy of Glenn Howe Quantitative traits are polygenic 55 Students at Connecticut Agricultural College, 1914 60 65 70 75 As the number of loci controlling a trait increases, the distribution of trait values in a population becomes bell-shaped 80 85 Influence of Environment on Human Height Mean = 67 2.7 in. 1914 Height vs GDP (1925-1949) By Country Mean = 70 3 in. 1996 Baten 2006 4:10 Schilling et al. 2002. Amer. Stat. 56: 223-229 6:5 Hartl and Clark 2007 Hartl, D. 1987. A primer of Population Genetics. 3 loci, 2 additive alleles Uppercase alleles contribute 1 unit to phenotype (e.g., shade of color) The phenotype is the outward manifestation of the genotype = + Phenotype Genotype σ2 P σ2G Environment σ2E Courtesy of Glenn Howe Types of genetic variance (σ2G) Additive (σ2A): effects of individual alleles Dominance locus Interaction (epistasis) (σ2D): effects of allele interactions within (σ2I): effects of interactions among loci σ2G = σ2A + σ2D + σ2I Non-additive Main cause for resemblance between relatives Heritability Phenotype vs Genotype Var(phenotype) = Var(genotype) + Var(environment) Heritability: Var(genotype) / Var(phenotype) Two types of heritability Broad-Sense Heritability includes all genetic effects: dominance, epistasis, and additivity − For example, the degree to which clones or monozygotic twins have the same phenotype Narrow-Sense Heritability includes only additive effects − For example, degree to which offspring resemble their parents Heritability (continued) Characteristic of a trait measured in a particular population in a particular environment Best estimated in experiments (controlled environments) Estimated from resemblance between relatives The higher the heritability, the better the prediction of genotype from phenotype (and vice versa) h² = 0.5 h² = 0.1 P h² = 0.9 P G P G http://psych.colorado.edu/~carey/hgss/hgssapplets/heritability/heritability1/heritability1.html G Identifying Genes Underlying Quantitative Traits Many individual loci are responsible for quantitative traits, even those with high heritability Identification of these loci is a major goal of breeding programs Allows mechanistic understanding of adaptive variation Methods usually rely on correlations between molecular marker polymorphisms and phenotypes Quantitative Trait Locus Mapping A B C A B C Parent 2 a b c X HEIGHT Parent 1 a b c A B C A B c B b a b c Bb A B C X A A b B c c a a BB c c BB F1 F1 BB a A B b c c bb modified from D. Neale A a b b c c bb AA bB cC BB bb a b c A A B b c c Bb a a B B c c Bb GENOTYPE A a b B c c Bb a B c BB BB Quantitative Trait Locus Analysis Step 1: Make a controlled cross to create a large family (or a collection of families) Parents should differ for phenotypes of interest Segregation of trait in the progeny Step 2: Create a genetic map Large number of markers phenotyped for all progeny Step 3: Measure phenotypes Need phenotypes with high heritability Step 1: Construct Pedigree Cross two individuals with contrasting characteristics Create population with segregating traits Ideally: inbred parents crossed to produce F1s, which are intercrossed to produce F2s Recombinant Inbred Lines created by repeated intercrossing Allows precise phenotyping, isolation of allelic effects Grisel 2000 Alchohol Research & Health 24:169 Step 2: Construct Genetic Map Number of recombinations between markers is a function of map distance Gives overview of structure of entire genome Anonymous markers are cheap and efficient: AFLP, Genotyping by Sequencing Codominant markers much more informative: SSR, SNP Genotyping by Sequencing gives best of both worlds: cheap, abundant, codominant markers! Step 3: Determine Phenotypes of Offspring 0.1 Phenotype must be segregating in pedigree Must differentiate genotype and environment effects How? 0.5 0.9 Works best with phenotypes with high heritability Step 4: Detect Associations between Markers and Single-marker associations are Phenotypes simplest Simple ANOVA, correcting for multiple comparisons Log likelihood ratio: LOD (Log10 of odds) LOD = log10 Pr(Data | QTL) Pr(Data | noQTL) If QTL is between two markers, situation more complex Recombination between QTL and markers (genotype doesn't predict phenotype) 'Ghost' QTL due to adjacent QTL Use interval mapping or composite interval mapping Simultaneously consider pairs of loci across the genome Step 5: Identify underlying molecular mechanisms QTL chromosome Genetic Marker QTG: Quantitative Trait Gene QTN: Quantitative Trait Nucleotide Adapted from Richard Mott, Wellcome Trust Center for Human Genetics QTL Limitations Huge regions of genome underly QTL, usually hundreds of genes How to distinguish among candidates? Biased toward detection of large-effect loci Need very large pedigrees to do this properly Limited genetic base: QTL may only apply to the two individuals in the cross! Genotype x Environment interactions rampant: some QTL only appear in certain environments Linkage Disequilibrium and Quantitative Trait Mapping Linkage and quantitative trait locus (QTL) analysis Need a pedigree and moderate number of molecular markers Very large regions of chromosomes represented by markers Association Studies with Natural Populations No pedigree required Need large numbers of genetic markers Small chromosomal segments can be localized Many more markers are required than in traditional QTL analysis Cardon and Bell 2001, Nat. Rev. Genet. 2: 91-99 ancestral chromosomes G T HEIGHT Association Mapping * TT TC GENOTYPE CC recombination through evolutionary history present-day chromosomes in natural population G C G T A C A C * G T A T * * Slide courtesy of Dave Neale Next-Generation Sequencing and Whole Genome Scans The $1000 genome is on the horizon Current cost with Illumina HiSeq 2000 is about $2000 for 10X depth Thousands of human genomes have now been sequenced at low depth Can detect most polymorphisms with frequency >0.01 True whole genome association studies now possible at a very large scale Direct to Consumer Genomics: 23 & Me and other genotyping services http://www.1000genomes.org/ Commercial Services for Human Genome-Wide SNP Characterization NATURE|Vol 437|27 October 2005 Assay 1.2 million “tag SNPs” scattered across genome using Illumina BeadArray technology Ancestry analyses and disease/behavioral susceptibility Identifying genetic mechanisms of simple vs. complex diseases Simple (Mendelian) diseases: Caused by a single major gene High heritability; often can be recognized in pedigrees Example: Huntington’s, Achondroplasia, Cystic fibrosis, Sickle Cell Anemia Tools: Linkage analysis, positional cloning Over 2900 disease-causing genes have been identified thus far: Human Gene Mutation Database: www.hgmd.cf.ac.uk Complex (non-Mendelian) diseases: Caused by the interaction between environmental factors and multiple genes with minor effects Interactions between genes, Low heritability Example: Heart disease, Type II diabetes, Cancer, Asthma Tools: Association mapping, SNPs !! Over 35,000 SNP associations have been identified thus far: http://www.snpedia.com Slide adapted from Kermit Ritland Complicating factor: Trait Heterogeneity Same phenotype has multiple genetic mechanisms underlying it Slide adapted from Kermit Ritland Case-Control Example: Diabetes Knowler et al. (1988) collected data on 4920 Pima and Papago Native American populations in Southwestern United States High rate of Type II diabetes in these populations Found significant associations with Immunoglobin G marker (Gm) Does this indicate underlying mechanisms of disease? Knowler et al. (1988) Am. J. Hum. Genet. 43: 520 Case-control test for association (case=diabetic, control=not diabetic) Gm Haplotype Type 2 Diabetes present absent Total present 8 29 37 absent 92 71 163 100 100 200 Total Question: Is the Gm haplotype associated with risk of Type 2 diabetes??? (1) Test for an association C21 = (ad - bc)2N . (a+c)(b+d)(a+b)(c+d) = [(8x71)-(29x92)]2 (200) = 14.62 (100)(100)(37)(163) (2) Chi-square is significant. Therefore presence of GM haplotype seems to confer reduced occurence of diabetes Slide adapted from Kermit Ritland Case-control test for association (continued) Question: Is the Gm haplotype actually associated with risk of Type 2 diabetes??? The real story: Stratify by American Indian heritage 0 = little or no indian heritage; 8 = complete indian heritage Index of indian Heritage 0 4 8 Conclusion: Gm Haplotype Percent with diabetes Present 17.8 Absent 19.9 Present 28.3 Absent 28.8 Present 35.9 Absent 39.3 The Gm haplotype is NOT a risk factor for Type 2 diabetes, but is a marker of American Indian heritage Slide adapted from Kermit Ritland Population structure and spurious association Assume populations are historically isolated One has higher disease frequency by chance Unlinked loci are differentiated between populations also Unlinked loci show disease association when populations are lumped together Population with high disease frequency Gene flow barrier Population with low disease frequency Alleles at neutral locus Alleles causing susceptibility to disease Association Study Limitations Population structure: differences between cases and controls Genetic heterogeneity underlying trait Random error/false positives Inadequate genome coverage Poorly-estimated linkage disequilibrium Association Analysis with a Mixed Model effects of background SNPs phenotype (response variable) of individual i effect of target SNP Family effect (Kinship coefficient) Population Effect (e.g., Admixture coefficient from Structure or values of Principal Components) Implemented in the Tassel program (Wednesday in lab)