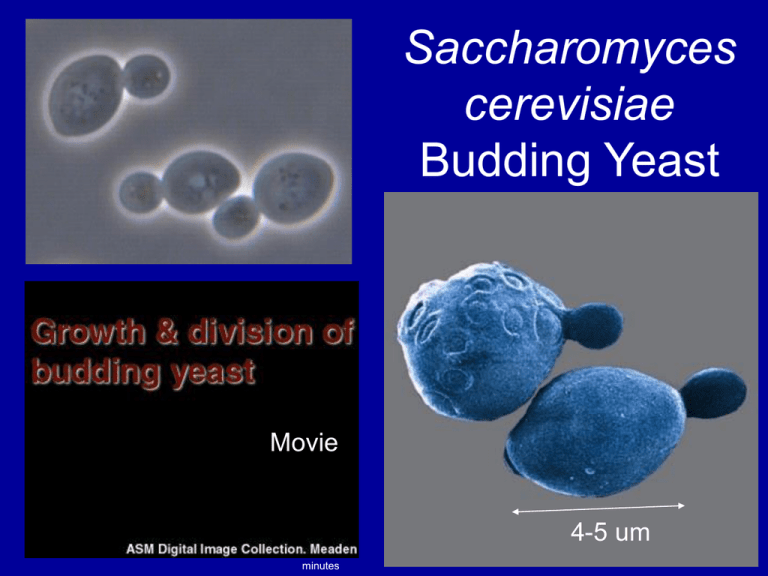
Saccharomyces
cerevisiae
Budding Yeast
Movie
4-5 um
minutes
Mitochondria
Bud site
EM view
Nucleolus
Vacuole
ER/
Golgi
Nucleus
Nuclear envelope
And
Nuclear Pores
Wall
~5 um
The Yeast Nuclear Genome
bp
16 Chromosomes (200-1600 Kbp)
12,052 Kbp nuclear genome (3.5x E. coli)
roughly 6000 protein-encoding genes
(the mitochondrial genome is about 76,000bp and encodes about 50 ORFs)
Transcriptome
Profiling
Human Disease Homologs
Yeast
Gene
TBLASTN
P-value
MSH2
3.8e-255
YCF1
2.4e-157
GEF1
3.4e-95
TEL1
8.8e-84
YNL161W 8.5e-82
SOD1
8.9e-56
SGS1
3.1e-50
IRA2
1.0e-28
Percent Percent
Identity Similarity
43
31
33
49
41
55
24
21
65
57
58
36
65
69
34
45
Human Gene ID
Mutator gene (MSH2, colon cancer)
Cystic fibrosis conductance regulator (CFTR)
Voltage-gated chloride ion channel
Ataxia telangiectasia gene
Myotonic dystrophy associated protein kinase
Superoxide dismutase (SOD-1)
Werner's Syndrome gene
Neurofibromin (NF1)
The Domestication of Yeast
The Yeast Life Cycle
“sex and the single yeast”
Diploid (2n)
Haploid (n)
Alternation of Generations
Mating
(Conjugation)
heterothallic
in the mother
cell mother cell
The Mitotic Cell Cycle
Master Regulatory Molecules
Control the Cycle
Transcription
factors
Cyclins (9 total) + CDK (Cdc28)
90 minute generation time
Ori Licensing
Controlling
S
Start
Bud Site Selection by Cdc42
A Cdc42ts
Mutant is
Defective in
Budding
and
Division
Superposition of
Spc42::GFP
to mark MTOCs
and Septin:GFP
to mark bud site
Septin::GFP
Septins Ring
the Bud Site
Cortical
Patches Target
Exocytosis
Contractile
Ring at
Cytokinesis
Long Cables
Support
Transport
Microfilaments
Target the Bud
The Secretory
Pathway
Delivers
Localized Wall
Synthesis
Controls
Growth
QuickTime™ and a
Cinepak Codec by Radius decompressor
are needed to see this picture.
Buds
Movie
An Intra-Nuclear
Spindle
Spc42::GFP
DNA
The Spindle Pole Body
The Fungal MTOC
The Cytoskeleton in Mitosis
QuickTime™ and a
Cinepak Codec by Radius decompressor
are needed to see this picture.
Tubulin::GFP
QuickTime™ and a
None decompressor
are needed to see this picture.
Myosin1(MyosinII)::GFP
homotypic mutants can’t switch
Mothers
Switch
in G1
A
A
HO
Alpha
A
HO
A
HO
Alpha
Alpha
Mating Type Switching
After Cytokinesis
A
alpha
a
a
Silent
HML
Active
MAT
Silent
HMR
alpha
alpha
a
alpha
a
a
Silenced
Switching is a
Gene Conversion
Event
Recombination Enhancer
Determines the Polarity of
the Switch
Silenced
Switching
Switching at MAT Changes
the Mating Type of the Cell
Certain mRNAs and proteins are
Transported into the Bud
QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture.
Myosin V
Ash1 protein
HO endonuclease
Ash1 mRNA encodes a
transcriptional repressor of HO
expression, suppressing
switching in the daughter cell
QuickTime™ and a
Graphics decompressor
are needed to see this picture.
Mating also
involves polarized
growth
Movie
Cdc42
CDC42 G-protein
FUS proteins
Pheromone
response
MAPKKKK
MAPKKK
MAPKK
MAPK
Ste5 is a
scaffolding
protein
responsible for
organizing a
MAP kinase
cascade
Candida albicans
Hyphal Growth
(Invasive)
A Few Questions for Thought
• Describe the life cycle of Saccharomyces cerevisiae,
noting relationships between mitotic growth, the alternation
of generations, meiosis, conjugation and spore formation.
•Review the compartments and cytoskeleton of the
eukaryotic cell, comparing and contrasting what you
learned in the first semester with this model yeast.
•Compare and contrast (in a simple way) mammalian and
fungal cell cycles.
•Describe the role(s) of septins, spindle pole bodies, and
CDC42 protein in the growth of E. coli.
•How is a cell’s mating type identity determined? How can
it switch (and why)? How is switching repressed in certain
cells? Why is this important?
•(After lab) Explain how a cell senses the presence of a
mating partner, and what happens once it shows interest?












