PowerPoint Presentation - AGRI-MIS
advertisement

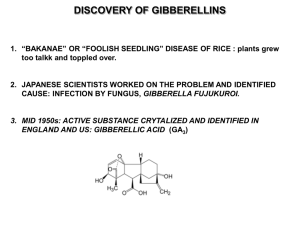
Gibberellins GAs Gibberellins (GAs) a class of plant hormones affect several important plant processes eg., seed germination stem elongation flowering male sterility Gibberellins 1926 Japanese scientist Gibberella fujikuroi gibberellin A (terpenoid cpd) 1954, 1955 US and UK scientists 1958 GA1 in higher plant GAx 1987 synthesis/metabolism Gibberellins 1991 84 GAs 1995 89 GAs 64 plants, 12 fungi 13 both 1996 more than 100 / 136 1997 genes being cloned Gibberellic acid (GA3) End metabolic product in fungi Plant GA20 GA5 GA3 Commercial High activity Slow degradation Similar to GA1 additional double bond Gibberellins GA4 GA7 nonpolar, slowly diffuse GA9 GA12 precursor GA29 GA34 deactivated form Different tissues Different forms of GA Gibberellins fungi algae bacteria moss fern gymnosperm angiosperm Gibberellins growing, differentiated tissues young, developing, expanding leaves developing seeds/fruit Gibberellins elongated internode/petiole shoot/stem apex root cap/tip xylem sap Synthesis and Metabolism Mevalonic acid pathway in cytosol Non mevalonic acid pathway in plastid Mevalonic acid pathway In higher plants from GA12 aldehyde Early 13-hydroxylation pathway (GA1) Non 13-hydroxylation pathway (GA4) with GA20oxidase genes: pathway shifted GA4 increased / GA1 decreased GA12 aldehyde: precursor of GA derivatives by oxidation (C20) and hydroxylation (C13 C3 C2) Vegetative tissue: conserved synthetic pathway 13-OH pathway to GA20 (C19-GA) then 3b-OH to GA1 except: arabidopsis and cucumber non 13-OH pathway to GA4 Reproductive tissue/seed: various pathways different forms of GA From mevalonic acid (6C) GGPP (20C-linear cpd) ent kaurene (1st specific cpd) GA12 aldehyde (first GA) GAx Gibberellins Isoprene (5C) as basic unit ent-Gibberellane skeleton tetracyclic diterpenoid cpd 2 main types: C20-GA and C19-GA GA derivatives by modification of 4 rings * C20 oxidation: CH3 CH2OH CHO COOH * Hydroxylation at C2 C3 and C13: number, position stoichiometry * Loss of C20 (C20 to C19 GA) GA inactivation * 2b-OH: GA20 GA1 * C20 oxidation GA29 GA8 to COOH GA inactivation * Conjugation by glucose Glycosylation: inactive, storage and transport Glucose via COOH: GA glycoside Glucose via OH: GA glycosyl ether GA synthesis mutants Pea na mutant: dwarf ent-kaurene Pea le mutant: dwarf GA12 aldehyde exogenous GA1 tall exogenous GA20 no response cloned Le gene: 3b hydroxylase GA20 GA1 Considering 2 loci na Le normal ent-kaurene Na le normal GA20 Grafting 1. na Le Na le 2. Na le Na Le Conclusion? scion stock tall scion stock dwarf GA mechanism in elongation Unlike auxin (acidification) Increase wall extensibility Decrease minimum force for wall extension GA mechanism in elongation By (may) decrease Ca concentration in the wall increase Ca uptake into the cell reduce crosslinking of lignin-related cpd (via peroxidase) GA mechanism in germination Activate transcription of a amylase gene In scutellum and aleurone GA detection and assay Bioassay Easy but not specific Fractionation Plant response Lettuce hypocotyls elongation Microdrop/dwarf rice a amylase production GC-MS Solvent extraction Chromatography (polarity) GC (boiling point) MS (mass) Identification and quantification High sensitivity and more specific GA inhibitors Inhibit ent-kaurene synthesis AMO1618 Cycocel Inhibit ent-kaurene oxidation Paclobutrazol Uniconazol Ancymidol Tetcyclasis Inhibit later steps by dioxygenases Bx-1112 LAB1988999 Hormone Responses Perception: receptor Signal transduction: second messenger (cAMP, cGMP) G protein Ca-Calmodulin enzyme transcription factor At last step Gene expression Specific region in promoter cis element DNA-binding protein transcription factor GA studies Exogenous GA / GA inhibitor GA mutant Gene identification / Gene cloning Gene expression / Transformation GA synthesis Enzyme: gene product of multigene family Each gene with specific pattern of expression AtGA20ox1: shoot growth AtGA20ox2: inflorescence development AtGA20ox3: early seedling development At later steps of synthetic pathway Genes controlled by GA, light and daylength GA: inhibit transcription of GA20oxidase (GA19 to GA20) inhibit 3b hydroxylase promote 2b hydroxylase Negative Light: feedback: promote reduce conversion production GA1 of active to inactive GA20GA8 and GA1 Daylength (LD): floralofinitiation activates activity reducing shoot GA20oxidase elongation GA53 to GA44 GA19 to GA20 Pea, Pisum sativum In de-etiolated pea seedling, exposed to red, blue, far red, all reduce GA1 level Lettuce: Lactuca sativa seed germination Red light: activates LsGA3ox1 expression GA1 increase Far-red light: inhibits LsGA3ox1 Auxin: promote GA1 production inhibit deactivation steps to GA29 and GA8 GA synthetic mutants Arabidopsis: seed germination assay 5 complementation groups (56 lines) ga1 ga2 ga3 ga4 and ga5 all recessive, dwarf, and male sterile ga1 and ga2 reversed by ent-kaurene ga3 reversed by ent-kaurenal Genes GA1 kaurene synthase (ent-CDP synthase) GA3 Cyt P450-dependent monooxygenase GA4 3b hydroxylase GA5 GA20oxidase Pea (sln) decrease 2b hydroxylase activity increase active GA tall plant with light green leaves Signal transduction mutants Stature mutants Decreased response to GA Increased response to GA Decreased signaling mutants Dwarf Complete phenocopy of GA-deficient mutants No response to exogenous GA Decreased signaling mutants Partially / fully dominant Arabidopsis gai Maize D8 D9 Wheat Rht1 Rht2 Rht3 Negative regulators Arabidopsis gai mutant Dwarf Higher level of active GA and GA20oxidase Semidominant Arabidopsis gai mutant gai1-1 51 bp inframe deletion loss of 17 amino acid constitutive repressor Arabidopsis gai mutant intragenic suppressor of gai loss of function allele WT phenotype Maize D8 mutant Dwarf Higher level of active GA 6 dominant alleles with different severity Wheat Rht mutant 8 dominant alleles with different severity Dwarf: prevent lodging Wheat + N fertilizer: increase yield increase height Norin10: dwarf line 2 mutated loci: Rht1 or Rht-B1b (chrs 4B) Rht2 or Rht-D1b (chrs 4D) All genes cloned: deduced amino acid sequence GAI / Rht / d8 homologs Conserved domains I and II in N terminal gai mutant: deletion in domain I D8 / Rht: mutation in domain I and/or II *N terminal essential for GA response* Increased signal transduction mutants Similar to WT + GA Tall by elongated internodes Arabidopsis Barley Rice Tomato Pea spy rga sln spy slr pro la crys Recessive / Negative regulators Increased signal transduction mutants Arabidopsis rga Identified by suppression analysis of ga1-3 New mutant: taller ga1-3 < ga1-3* < WT new locus: repressor of ga1-3 (rga) Increased signal transduction mutants rga: recessive (deletion mutation) increase stem elongation reverse ga1-3 delayed flowering time no effect on GA biosynthesis RGA: negative regulator Gene: 82% homology to GAI especially in N region Original gai mutant: gain of function Loss of function allele of GAI ? Phenotype: normal Increase paclobutrazol resistance Low GA = normal height At least two components in Arabidopsis GA signaling pathway GAI and RGA homopolymeric Serine / Threonine residue leucine heptad for protein-protein interaction putative nuclear localizing signal Barley sln slender mutant recessive long internodes and narrow leaves male sterile increase a-amylase w/o GA low endogenous GA resistant to GA synthesis inhibitors negative regulator sln x dwarf mutant = sln phenotype SLN = GAI/RGA homolog Dominant allele of SLN mutant Mutation in N terminal Dwarf barley Rice slr slender rice recessive phenocopy of barley sln 1 bp deletion in NLS domain (nuclear localization signal ) Rice slr frame shift mutation stop codon truncated protein SLR gene = SLN homolog Modified SLR: 17 aa deletion in DELLA domain Transformation: dwarf rice GA signal component Dicot / Monocot GAI RGA Rht d8 SLN SLR Putative transcription repressor Arabidopsis spy spindly mutant, recessive paclobutrazol-resistant long hypocotyls light green leaves early flowering spy ga1-2 = spy phenotypes spy gai = spy phenotypes Arabidopsis spy SPY gene product: O-GlcNAc transferase Signaling molecule Involved in protein-protein interaction Negative regulator Before responses Expression of GA-regulated genes: Protein-DNA interaction Transcription factor cis elements Transcription factor: GAMyb Barley: HvGAMyb Bind specific sequence in promoter of a-amylase gene Increase gene expression Overexpression of HvGAMyb gene = GA treatment Arabidopsis: GAMyb-like genes AtMyb33 AtMyb65 AtMyb101 Functional homologs of barley GAMyb Transform barley aleurone with AtMyb33 Activate a-amylase production Arabidopsis: facultative LD plants Transfer plants from SD to LD 11x increase of GA1 3x increase of GA4 increase AtMyb33 expression in shoot apex shoot apex transition to flowering Potential target for AtMyb LFY promoter LEAFY: meristem-identity gene Evidence AtMyb binding to a specific 8-bp sequence in LFY promoter cis elements specific regions in promoter transcription factor binding site identified by deletion or site specific mutagenesis: gene expression after promoter modification Conserved sequences among GA-regulated genes - amylase box: TATCCAT - GARE: TAACAA/GA - Pyrimidine box: C/TCTTTTAC/T GA and a-amylase production Perception at membrane receptors Increase intracellular Ca Decrease intracellular pH Increase [CaM] Increase cGMP Increase GAMyb transcription Increase a-amylase activity Some protein phosphorylation