ppt - University of Connecticut
advertisement
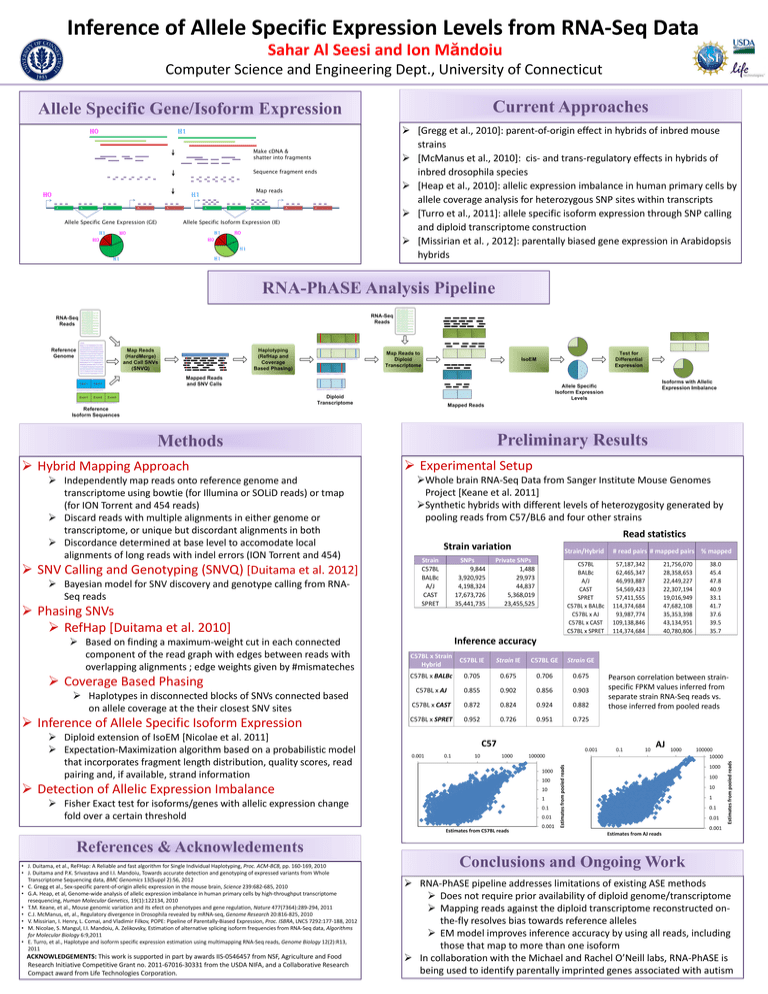
Inference of Allele Specific Expression Levels from RNA-Seq Data Sahar Al Seesi and Ion Măndoiu Computer Science and Engineering Dept., University of Connecticut Current Approaches Allele Specific Gene/Isoform Expression H0 H1 Make cDNA & shatter into fragments Sequence fragment ends H0 Map reads H1 A B C D Allele Specific Gene Expression (GE) H1 E A B C D E Allele Specific Isoform Expression (IE) H1 H0 H0 H0 H0 H1 H1 H1 [Gregg et al., 2010]: parent-of-origin effect in hybrids of inbred mouse strains [McManus et al., 2010]: cis- and trans-regulatory effects in hybrids of inbred drosophila species [Heap et al., 2010]: allelic expression imbalance in human primary cells by allele coverage analysis for heterozygous SNP sites within transcripts [Turro et al., 2011]: allele specific isoform expression through SNP calling and diploid transcriptome construction [Missirian et al. , 2012]: parentally biased gene expression in Arabidopsis hybrids RNA-PhASE Analysis Pipeline Preliminary Results Methods Hybrid Mapping Approach Independently map reads onto reference genome and transcriptome using bowtie (for Illumina or SOLiD reads) or tmap (for ION Torrent and 454 reads) Discard reads with multiple alignments in either genome or transcriptome, or unique but discordant alignments in both Discordance determined at base level to accomodate local alignments of long reads with indel errors (ION Torrent and 454) SNV Calling and Genotyping (SNVQ) [Duitama et al. 2012] Bayesian model for SNV discovery and genotype calling from RNASeq reads Phasing SNVs RefHap [Duitama et al. 2010] Based on finding a maximum-weight cut in each connected component of the read graph with edges between reads with overlapping alignments ; edge weights given by #mismateches Coverage Based Phasing Haplotypes in disconnected blocks of SNVs connected based on allele coverage at the their closest SNV sites Inference of Allele Specific Isoform Expression Experimental Setup Whole brain RNA-Seq Data from Sanger Institute Mouse Genomes Project [Keane et al. 2011] Synthetic hybrids with different levels of heterozygosity generated by pooling reads from C57/BL6 and four other strains Read statistics Strain variation Strain C57BL BALBc A/J CAST SPRET SNPs 9,844 3,920,925 4,198,324 17,673,726 35,441,735 Strain/Hybrid Private SNPs 1,488 29,973 44,837 5,368,019 23,455,525 C57BL BALBc A/J CAST SPRET C57BL x BALBc C57BL x AJ C57BL x CAST C57BL x SPRET # read pairs # mapped pairs % mapped 57,187,342 62,465,347 46,993,887 54,569,423 57,411,555 114,374,684 93,987,774 109,138,846 114,374,684 21,756,070 28,358,653 22,449,227 22,307,194 19,016,949 47,682,108 35,353,398 43,134,951 40,780,806 38.0 45.4 47.8 40.9 33.1 41.7 37.6 39.5 35.7 Inference accuracy C57BL x Strain Hybrid C57BL IE Strain IE C57BL GE Strain GE C57BL x BALBc 0.705 0.675 0.706 0.675 C57BL x AJ 0.855 0.902 0.856 0.903 C57BL x CAST 0.872 0.824 0.924 0.882 C57BL x SPRET 0.952 0.726 0.951 0.725 Pearson correlation between strainspecific FPKM values inferred from separate strain RNA-Seq reads vs. those inferred from pooled reads Diploid extension of IsoEM [Nicolae et al. 2011] Expectation-Maximization algorithm based on a probabilistic model that incorporates fragment length distribution, quality scores, read pairing and, if available, strand information Detection of Allelic Expression Imbalance Fisher Exact test for isoforms/genes with allelic expression change fold over a certain threshold References & Acknowledements • J. Duitama, et al., ReFHap: A Reliable and fast algorithm for Single Individual Haplotyping, Proc. ACM-BCB, pp. 160-169, 2010 • J. Duitama and P.K. Srivastava and I.I. Mandoiu, Towards accurate detection and genotyping of expressed variants from Whole Transcriptome Sequencing data, BMC Genomics 13(Suppl 2):S6, 2012 • C. Gregg et al., Sex-specific parent-of-origin allelic expression in the mouse brain, Science 239:682-685, 2010 • G.A. Heap, et al, Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing, Human Molecular Genetics, 19(1):122134, 2010 • T.M. Keane, et al., Mouse genomic variation and its efect on phenotypes and gene regulation, Nature 477(7364):289-294, 2011 • C.J. McManus, et, al., Regulatory divergence in Drosophila revealed by mRNA-seq, Genome Research 20:816-825, 2010 • V. Missirian, I. Henry, L. Comai, and Vladimir Filkov, POPE: Pipeline of Parentally-Biased Expression, Proc. ISBRA, LNCS 7292:177-188, 2012 • M. Nicolae, S. Mangul, I.I. Mandoiu, A. Zelikovsky, Estimation of alternative splicing isoform frequencies from RNA-Seq data, Algorithms for Molecular Biology 6:9,2011 • E. Turro, et al., Haplotype and isoform specific expression estimation using multimapping RNA-Seq reads, Genome Biology 12(2):R13, 2011 ACKNOWLEDGEMENTS: This work is supported in part by awards IIS-0546457 from NSF, Agriculture and Food Research Initiative Competitive Grant no. 2011-67016-30331 from the USDA NIFA, and a Collaborative Research Compact award from Life Technologies Corporation. Conclusions and Ongoing Work RNA-PhASE pipeline addresses limitations of existing ASE methods Does not require prior availability of diploid genome/transcriptome Mapping reads against the diploid transcriptome reconstructed onthe-fly resolves bias towards reference alleles EM model improves inference accuracy by using all reads, including those that map to more than one isoform In collaboration with the Michael and Rachel O’Neill labs, RNA-PhASE is being used to identify parentally imprinted genes associated with autism