The Secretory Pathway
advertisement

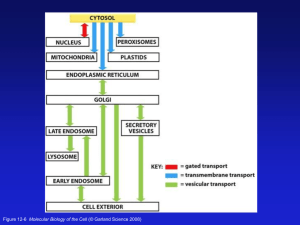
GBS709: Biological Organization- Secretion David M. Bedwell, Ph.D. Office: BBRB432 Phone: 934-6593 E-mail: dbedwell@uab.edu Reading: Alberts et al., Molecular Biology of the Cell (5th Ed.): Endoplasmic Reticulum: Chapter 12, pp. 723-745 Vesicular Traffic: Chapter 13, pp. 749-787 Major Intracellular Compartments of an Animal Cell Figure 12-1 Molecular Biology of the Cell (© Garland Science 2008) Topological Relationships Between Compartments of the Secretory Pathway Figure 12-5 Molecular Biology of the Cell (© Garland Science 2008) Simplified Roadmap of Intracellular Protein Traffic Figure 12-6 Molecular Biology of the Cell (© Garland Science 2008) Table 12-3 Molecular Biology of the Cell (© Garland Science 2008) General Features of the ER The endoplasmic reticulum (ER) is a highly convoluted, single membrane-bound organelle that contains as much as 50% of the total membrane of liver cells. Due to tubular nature of the ER, the total membrane surface area of ER in liver cells is ~25 times the surface area of the plasma membrane. The ER membrane is thought to enclose a single internal space called the lumen, which can comprise >10% of the total cell volume. The ER Frequently Exhibits a Reticular Structure Fluorescent visualization of the endoplasmic reticulum (ER) showing it frequently exists as a tubular network throughout the cytosol. The Reticular Portion of the ER Is a Highly Dynamic Tubular Network Rough and Smooth ER The smooth ER (SER) is the site of most lipid biosynthesis in the cell. The rough ER (RER) is the site of entry of proteins into the secretory pathway, and is so named because ribosomes located on the rough ER (carrying out the translocation of proteins into the ER) give this membrane a "rough" appearance when viewed under the microscope. Transitional elements (part rough and part smooth) are the site at which transport vesicles bud off to carry newly synthesized lipids and proteins to the Golgi apparatus. Rough and Smooth ER Rough and smooth ER in a liver cell Figure 12-36 Molecular Biology of the Cell (© Garland Science 2008) Isolation of Rough and Smooth Microsomes from the ER Figure 12-37 Molecular Biology of the Cell (© Garland Science 2008) The Signal Hypothesis [Blobel and Dobberstein, J. Cell Biol. 67: 835 (1975)] Gunter Blobel won the 1999 Nobel Prize for discovering the mechanism of ER protein targeting. Figure 12-38 Molecular Biology of the Cell (© Garland Science 2008) Evidence for a Proteinaceous Channel Rough microsomes contain channels of high ion conductivity that can be activated by puromycin, an antibiotic that causes the release of nascent chains from ribosomes. This suggested that when the nascent chain is released, it leaves a "hole" (the postulated protein conducting channel) in the ER membrane. This channel remained open until the membranes were washed with high salt, which caused the ribosomes to dissociate into their subunits. These results are most consistent with a model in which a protein-lined channel in the ER membrane opens when a ribosome binds to the membrane and resumes translocation. When the ribosome is tricked into releasing the nascent chain prematurely with puromycin, the channel remains open (thus allowing ions to flow through) until the ribosome dissociates, which causes the channel to close. Overview of ER Translocation The translocation of proteins into the ER is generally a cotranslational process and can be divided into two phases: targeting of the nascent polypeptide to the cytoplasmic surface of the ER membrane. translocation of the polypeptide across the ER membrane. Targeting Secretory Proteins to the ER In eukaryotic cells, many proteins are targeted to the ER by a ribonucleoprotein complex called the Signal Recognition Particle (SRP). Targeting occurs in a co-translational manner. Once ~30 amino acids of the nascent chain emerge from the ribosome, it is bound by the SRP and translation is temporarily arrested (this represents ~ 70 amino acids total, since 30-40 amino acids are buried within the ribosome). Following the translational arrest, the SRP-ribosome complex moves to the ER membrane where SRP docks to a specific protein complex on the ER surface called the SRP receptor (the ribosome may also associate with a ribosome receptor on the ER membrane). The translational arrest is relieved when the SRP releases and recycles for another round, allowing translation of the ER bound secretory protein to resume. Signal Recognition Particle (SRP) (Binds SRP Receptor) SRP is an 11S ribonucleoprotein (RNP) complex composed of a 7S RNA (~300 nucleotides) and six polypeptide chains. Signal Recognition Particle (SRP) The SRP contains three functional domains: The p19/54 domain binds to the signal sequence. p54 binds the signal sequence, while p19 links p54 to the 7S RNA. The p9/14 domain binds to the A site of the ribosome and arrests translation elongation. The p68/72 domain helps initiate insertion of the signal sequence into the ER membrane by binding to the SRP receptor. Signal Recognition Particle (SRP) Mammalian SRP Figure 12-39 Molecular Biology of the Cell (© Garland Science 2008) SRP bound to a ribosome How an ER Signal and SRP Direct Ribosomes to the ER Figure 12-40 Molecular Biology of the Cell (© Garland Science 2008) Pools of Free and Membrane Bound Ribosomes Figure 12-41 Molecular Biology of the Cell (© Garland Science 2008) Structure of the Sec61 Complex Figure 12-42 Molecular Biology of the Cell (© Garland Science 2008) Ribosome Bound to the Eukaryotic ER Protein Translocator Figure 12-43 Molecular Biology of the Cell (© Garland Science 2008) Cryo-EM Reconstruction Movie of the Yeast Ribosome and ER Translocator Components of the ER Translocase- Sec61p Sec61p has been found to interact directly with translocating polypeptides by crosslinking experiments in yeast and mammalian cells (the yeast and mammalian forms of Sec61p are 56% identical). Sec61p also interacts directly with ribosomes, and also functions as a ribosome receptor. Crosslinking of Sec61p to translocating precursors occurs in the ATP-dependent phase of the transport reaction, indicating that the interaction occurs while the precursor is moving across the membrane. Interestingly, Sec61p is a polytopic membrane protein that contains 10 membrane spanning domains, and shares structural and amino acid sequence homology with the E. coli SecY protein. Components of the ER Translocase- BiP BiP stands for Immunoglobulin Heavy Chain Binding Protein, and is called Kar2p in yeast. BiP is a resident ER protein that is related to the Hsp70 class of proteins and is thought to bind aggregated or misfolded proteins to prevent them from leaving the ER. It may also help refold them using an ATP dependent activity. BiP can also be crosslinked to translocating precursors in yeast. This result is also consistent with studies where it was found that mutationally altered Kar2p blocked not only precursor translocation, but also precursor interaction with Sec61p Peripheral Components of the ER Translocase Other proteins (Sec62, Sec63, Sec71 and Sec72) can assist with post-translational import. Sec63 contains a J domain and binds BiP. Signal peptidase- The signal peptidase has been purified as a complex from both mammalian and yeast microsomes, and each enzyme contains five different proteins. The catalytic site is on the trans (lumenal) side of the ER membrane. The subunits of this complex appear to span the membrane only once and have a second hydrophobic region that may interact with signal sequences. Oligosaccharyltransferase is the enzyme responsible for the transfer of an oligosaccharyl moiety to an asparagine residue in a consensus (Asn-X-Ser/Thr) recognition motif. The mammalian enzyme consists of three subunits. Neither signal peptidase or the oligosaccharyltransferase are required for protein translocation. 3 Ways for Protein Translocation Through Structurally Similar Translocators Figure 12-44 Molecular Biology of the Cell (© Garland Science 2008) Yeast Genetic Analysis Played a Key Role in Identifying Key Proteins in the Secretory Pathway Membrane Protein Insertion: Model For Soluble Protein Translocation Across the ER Membrane Figure 12-45 Molecular Biology of the Cell (© Garland Science 2008) Membrane Protein Insertion: Model For Integration of a Single-Pass Transmembrane Protein into the ER Membrane Figure 12-46 Molecular Biology of the Cell (© Garland Science 2008) Membrane Protein Insertion: Integration of SinglePass Transmembrane Proteins with an Internal Signal into the ER Membrane Figure 12-47 Molecular Biology of the Cell (© Garland Science 2008) Membrane Protein Insertion: Integration of a DoublePass Transmembrane Protein with an Internal Signal into the ER Membrane Figure 12-48 Molecular Biology of the Cell (© Garland Science 2008) Membrane Protein Insertion: Insertion of the Multipass Membrane Protein Rhodopsin into the ER Membrane Figure 12-49 Molecular Biology of the Cell (© Garland Science 2008) Asparagine-Linked (N-Linked) Precursor Oligosaccharides are Added to Proteins in the ER Figure 12-50 Molecular Biology of the Cell (© Garland Science 2008) Protein Glycosylation in the Rough ER Figure 12-51 Molecular Biology of the Cell (© Garland Science 2008) N-Linked Glycosylation Helps the ER Monitor Protein Folding Figure 12-53 Molecular Biology of the Cell (© Garland Science 2008) Export and Degradation of Misfolded ER Proteins via ER-Associated Decay (ERAD) Figure 12-54 Molecular Biology of the Cell (© Garland Science 2008) The Unfolded Protein Response (UPR) The unfolded protein response (UPR) results from the accumulation of misfolded proteins in the ER due to cellular or environmental stress. UPR triggers activation of a wide range of genes encoding proteins involved in protein folding and glycosylation in the ER, proteins involved in ER Associated Decay (ERAD), and proteins involved in vesicular transport. Thus, quality control, ERAD and UPR are tightly coordinated processes. 3 Parallel Signaling Pathways Activated by the Unfolded Protein Response (UPR) Figure 12-55a Molecular Biology of the Cell (© Garland Science 2008) Comparison of Mammalian and Yeast UPR Basic features of mammalian and yeast UPR are similar, but mammalian UPR has more “bells & whistles”. - Mammals & yeast: Ire1p sensor/kinase serves to splice the XBP1 mRNA, allowing the translation of the transcription factor Xbp1. - Mammals only: Another transcription factor, ATF6, is cleaved directly from the cytoplasmic domain of an ER transmembrane sensor. - Mammals only: Translational repression is mediated by another transmembrane sensor/kinase, PERK. Together, these responses to unfolded proteins in the ER allows mammalian cell to coordinately regulate transcription and translation to rapidly recover from ER stress. If the stress is too severe, apoptosis is induced. Transmembrane ER Stress Sensors That Regulate the UPR IRE1 splices XBP1 mRNA PERK blocks translation initiation ATF6 contains a transcription factor Regulated XBP1 mRNA Splicing is One Pathway Activated During the Unfolded Protein Response Ire1p XBP1 Xbp1 Figure 12-55b Molecular Biology of the Cell (© Garland Science 2008) Induction of Transcription During the UPR by XBP1 and ATF6 Regulation of Translation During the UPR eIF2 phosphorylation inhibits bulk translation. The presence of uORFs in certain mRNAs allows the translation of specific proteins during the UPR that turn on UPRresponsive genes. ER Questions? Proteins Transit From the ER to the Golgi Apparatus and Through the Secretory Pathway Occurs via Vesicular Transport Figure 13-3 Molecular Biology of the Cell (© Garland Science 2008) Basic Features of Vesicular Transport Figure 13-2 Molecular Biology of the Cell (© Garland Science 2008) 3 types of Vesicle Coat Proteins Figure 13-4 Molecular Biology of the Cell (© Garland Science 2008) Use of Different Coats in Vesicular Traffic Figure 13-5 Molecular Biology of the Cell (© Garland Science 2008) Different Phosphoinositides Mark Organelles and Membrane Domains Phosphotidylinositol (PI) and Phosphoinositides (PIPs) Figure 13-10 Molecular Biology of the Cell (© Garland Science 2008) Localized kinases and phosphatases in specific membranes regulate the production of PIPs which recruit binding proteins to specific membranes Intracellular Location of Phosphoinositides Figure 13-11 Molecular Biology of the Cell (© Garland Science 2008) Monomeric GTPases That Control Coat Assembly Monomeric GTPases Control Coat Assembly Arf proteins mediate COPI and clathrin coat assembly at Golgi membranes. Sar1 protein mediates COPII coat assembly at the ER membrane. Rab proteins guide vesicle targeting >60 members in Rab family. Each binds to specific membranes. Each Rab is associated with one or more membrane-enclosed organelles secretory or endocytic pathways. Each organelle has at least one Rab on its cytosolic surface. Soluble Rab-GDI (GDP dissociation inhibitor) keeps Rabs soluble and inactive. Membrane bound Rab-GEF (GTP Exchange Factor) activate Rabs via GTP-binding. Rab-GTP binds Rab effectors on membranes that mediate vesicular transport, membrane tethering and fusion. Formation of COPII-Coated Vesicles at the ER Membrane - Sar1 is a coat-recruitment GTPase. - Sar1-GEF is a GDP/GTP exchange factor. - Activated Sar1-GTP binds membrane via an amphipathic helix. - Sar1 binding initiates membrane curvature. Figure 13-13a/b Molecular Biology of the Cell (© Garland Science 2008) - Sar1 recruits a complex of COPII coat proteins (Sec23/24). - Sec24 binds cytoplasmic tails of cargo receptors. - COPII coat curves membrane of vesicle. Formation of COPII-Coated Vesicles at the ER Membrane - 2 additional COPII coat proteins, Sec13/31, forms the outer sell of the coat. - Sec13/31 can assemble into cages with the dimensions of COPII-coated vesicles. Figure 13-13c/d Molecular Biology of the Cell (© Garland Science 2008) - Membrane-bound, active Sar1-GTP recruits COPII subunits to the membrane. - This causes a membrane bud to form, which includes selected transmembrane proteins as cargo. - A subsequent membrane fusion event pinches off the coated vesicle. Tethering of a Vesicle to a Target Membrane Figure 13-14 Molecular Biology of the Cell (© Garland Science 2008) Structure of a Trans-SNARE Complex Figure 13-16 Molecular Biology of the Cell (© Garland Science 2008) Model of How SNARE Proteins Catalyze Membrane Fusion Figure 13-17 Molecular Biology of the Cell (© Garland Science 2008) NEM-Sensitive Factor (NSF) Dissociates SNARE Pairs After a Membrane Fusion Cycle N-Ethylmaleimide (NEM) is a thiol-reactive compound that inactivates NSF. Figure 13-18 Molecular Biology of the Cell (© Garland Science 2008) Recruitment of Cargo Molecules into ER Transport Vesicles A typical transport vesicle contains about 200 membrane proteins as cargo. Figure 13-20 Molecular Biology of the Cell (© Garland Science 2008) Retention of Incompletely Assembled Antibody Molecules in the ER Figure 13-21 Molecular Biology of the Cell (© Garland Science 2008) Homotypic Membrane Fusion Figure 13-22 Molecular Biology of the Cell (© Garland Science 2008) Vesicular Tubular Clusters Form Between the ER and the cis Golgi Network Figure 13-23a/b Molecular Biology of the Cell (© Garland Science 2008) Model for the Retrieval of Soluble ER Resident Proteins Figure 13-24a Molecular Biology of the Cell (© Garland Science 2008) The Golgi Apparatus Figure 13-25a/b Molecular Biology of the Cell (© Garland Science 2008) Oligosaccharide Processing in Golgi Compartments Figure 13-28 Molecular Biology of the Cell (© Garland Science 2008) 2 Classes of N-Linked Oligosaccharides Found in Mature Mammalian Glycoproteins Figure 13-30 Molecular Biology of the Cell (© Garland Science 2008) Oligosaccharide Processing in the ER and Golgi Apparatus Figure 13-31 Molecular Biology of the Cell (© Garland Science 2008) N- and O-Linked Glycosylation Figure 13-32 Molecular Biology of the Cell (© Garland Science 2008) What’s the Purpose of Glycosylation? Makes protein folding intermediates more soluble and prevents protein aggregation. Sequential modifications of N-linked sugars make a “glyco-code” that marks the progression of protein folding and mediates the binding of protein to chaperones and lectins (e.g., in ER quality control). Sugars have limited flexibility. This can limit the approach of other proteins (such as proteases) to the glycoprotein surface and reduce proteolysis. Extensive glycosylation of proteins (e.g., in the mucus coat of lungs and intestinal cells) protects against many pathogens. Recognition of glycoprotein sugar chains by lectins in the extracellular space is important for developmental processes and cell-cell recognition. 2 Models to Explain Golgi Organization and Protein Transport Between Cisternae Both mechanisms are probably used to maintain Golgi compartmental integrity Figure 13-35 Molecular Biology of the Cell (© Garland Science 2008) Golgi Questions?