Details of Ch. 17 Protein Synthesis PPT
advertisement

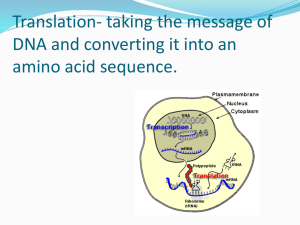
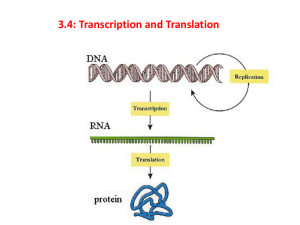
Protein Synthesis: From Gene to Protein Step #1 DNA unwinds and exposes the bases (Initiation) Begins at the Promoter Site How is a gene found from 3 billion base pairs? 1. TATA Box (25 bases upstream) a. Area that signals the RNA Polymerase II to attach to the DNA b. Transcription Factors bind to the TATA Box to help the RNA Polymerase II to attach to the DNA 2. Transcription Initiation Complex: Transcription Factors + RNA Polymerase at the promoter site Promoter Site TATAAAA ATATTTT Template DNA (Gene) TATA BOX Initial Transcription Factor Transcription Factors Transcription Initiation Complex Why is this important? RNA Polymerase II Step #2 mRNA made from DNA (Transcription) Promoter region Transcription unit (gene) Initiation: Covered in Step One RNA Polymerase Coding strand Elongation: RNA Polymerase moves downstream making RNA in the 5’ 3’ direction of the template strand mRNA Termination Sequence Termination: RNA Polymerase released when a terminator sequence is reached mRNA (AATAAA) RNA Polymerase Step #3 mRNA Leaves Nucleus and Goes to the Ribosome A. RNA Processing: mRNA is modified before it leaves the nucleus Pre-mRNA mRNA used in Translation Processing 1. Alteration at the ends of the pre-mRNA Leader Coding Segment Start Codon Trailer Stop Codon Termination Sequence Addition of the 5’ Cap Addition of the Poly (A) tail 1. Prevents degradation by enzymes 1. Prevents degradation by enzymes 2. Allows attachment to ribosome 2. Facilitates export from the nucleus 2. Alteration of the coding sequence - mRNA Splicing Pre-mRNA 5’ Cap Exon Intron Exon mRNA Exon Coding Segment Poly (A) tail Part of the RNA that remains to be translated into protein Intron Part of the RNA that is removed Intron 5’ Cap Poly (A) tail Enzymes 5’ Cap Exon Intron Exon 1 + Exon 2 + Exon 3 Coding Segment Poly (A) tail RNA Processing Vocabulary Pre-RNA Exon Intron Exon Proteins snRNA snRNA’s Small Nuclear RNA snRNP’s ‘SNURPS” Small Ribonucleoproteins Proteins snRNP’s Spliceosome Spliceosome Ribozymes The combination of snRNA + snRNP + Protein used to cut out introns RNA that function as enzymes. Some RNA’s can splice themselves What are the Functions of introns and exons? Spliceosome Components Removed Intron mRNA 1. Allows one code to make different proteins from generic “mix and match” domains 2. Controls gene expression B. Attachment to the Ribosome A Site 1. Ribosome Anatomy P site Large Subunit E Site Small Subunit 2. Steps of Attachment (Initiation) mRNA binding site a) mRNA attaches to the smaller unit b) Initiator tRNA with anticodon UAC bonds to start codon AUG c) Large ribosomal unit joins small unit with the tRNA at the P site. d) GTP provides the energy Met UA C A U G Met GTP Slide 9 Step #4 tRNA Bonds to Appropriate Amino Acid Specific Amino Acid Requirements Aminoacyl-tRNA Synthase 1. Aminoacyl-tRNA Synthase enzyme 2. Specific Amino Acid Active site fits specific Amino Acid 3. ATP 4. Specific tRNA tRNA Active Site specific for anticodon Aminoacyl tRNA “activated amino acid” Step # 5 tRNA bonds to mRNA at rRNA Step # 6 Amino Acids Bond to make the Protein All part of Translation Steps in Translation 1. Initiation – attachment of mRNA to the ribosome (This was already covered in Step # 3) 2. Elongation – the addition of amino acids to the growing protein chain A Site P site a) Codon Recognition : tRNA binds to codon at A site E Site GTP GDP GDP c) Translocation: tRNA in P site is shifted to the E site b) Peptide Bond Formation: Amino acids bond to form protein chain GTP Note: Growing Chain 3. Termination: Completed Protein and mRNA released from the Ribosome Release Factor Protein a) Release factor protein binds at stop codon at the A site Stop Codons (UAG, UAA, UGA) b) The release factor breaks bond of the tRNA and the last amino acid of the ribosome releasing the tRNA c) The 2 ribosomal units, mRNA, release factor dissociate Step #7 tRNA goes back to pick up more amino acids; mRNA breaks down; Funtional protein ready for use A. mRNA 1. May not immediately break down but bond to another ribosome to form a polyribosome 2. Many proteins can be made with one mRNA transcript B. Protein Most proteins must be modified to be functional -amino acids cleaved (methionine), functional groups added, tertiary and quaternary structures modified 1. Chaperonins needed to fold protein into correct tertiary structure 2. Secretory proteins are targeted to be modified in the Rough ER prior to export Reading the Code 3’ – TACGGCCGATTCTGACATCGAACT-5’ 5’ – ATGCCGGCTAAGACTGTAGCTTGA-3’ 3’ – TAC GGC CGA TTC TGA CAT CGA ACT – 5’ 5’ - ATG CCG GCT AAG ACT GTA GCT TGA – 3’ mRNA – AUG CCG GCU AAG ACU GUA GCU UGA (codons) tRNA – UAC GGC CGA UUC UGA CAU CGA ACU (anticodons) Amino Acids = Met – Pro – Ala – Lys – Thr – Val – Ala Mutations: Changes in the DNA Sequence Type of Mutation A. Chromosomal rearrangement (aneuploidy, trisomy, polyploidy) B. Changes in DNA base pairs (Point Mutations) Typical Effect of Mutation A. Usually lethal due to high number of genes (proteins) involved B. Effect Varies 1. Silent – no effect due to the redundancy of the triplet code 2. Missense – Little effect resulting with a functional protein 3. Nonsense – Protein nonfunctional C. Point Mutations Types 1. Base- Pair Substitutions Replacing one base pair with another 2. Deletions – removal of one base pair 3. Insertions – addition of one base pair Usually silent due to redundancy and “wobble” of code. Effect depends on: 1) Amino Acid type changed 2) Change in start or stop codons Cause frame shift mutations forming nonsense proteins THE END Slide 6 Slide 3 Slide 8 Ribosome Anatomy Growing Protein Chain Large Subunit Computer Model From X-Ray Crystallography Data E P A Small Subunit mRNA Slide 7 A Point Mutation with Bad Effects - Sickle Cell Anemia Difference in electronegativity of this amino acid causes hemoglobin to change shape after it gives up it’s O2 No Change due to redundancy of the code or “wobble” Different Amino Acid Effect depends on the amino acid type 1. Hydrophobic or hydrophlic 2. Positive or negative 3. Acidic or basic A premature stop codon will prevent from forming the complete protein THEREDDOGANDCAT Reading frame THE RED DOG AND CAT R Deletion frameshift THE EDD OGA NDC AT R Insertion frameshift THE RRE DDO GAN DCA T Genetic Code Redundancy 1. There are multiple codes for the same amino acids 2. The most variation is found in the 3rd base. “Wobble” 3. Some base pairs will bond to its “wrong” pair in the 3rd base 4. Some tRNA use the base Inosine(I) as their 3rd base that can bond to any base Slide 13 Slide 14