Fundamentals II:
Introduction to Bacteriology and
Bacterial Structure
Janet Yother, Ph.D.
Department of Microbiology
jyother@uab.edu
4-9531
Learning Objectives
• Fundamental properties of prokaryotes
• Basic structures of bacteria
• Gram-positive vs Gram-negative bacteria
Domains (Kingdoms)
Based on evolutionary relationships
• Eukaryote (Plants, Animals, Protists, Fungi)
• Eubacteria (Eubacteria)
• Archaea (Archaea)
Distinctive Features of Prokaryotic
and Eukaryotic Cells
Cell component
Prokaryotes
Eukaryotes
Nucleus
No membrane; single, (usually)
circular chromosome
Membrane-bound; a
number of individual
chromosomes
Extrachromosomal DNA Often present (plasmids, phage)
In organelles
Organelles in Cytoplasm None
Mitochondria (and
chloroplasts in
photosynthetic organisms)
Cytoplasmic Membrane
Respiration, secretion,
macromolecular synthesis
Lacks functions of
prokaryotic membrane
Cell Wall
Peptidoglycan (absent in
Mycoplasma)
No peptidoglycan
(cellulose, chitin in some)
Sterols
Absent (except in Mycoplasma)
Usually present
Ribosomes
70S (50S + 30S)
80S (60S + 40S)
Bacterial Nomenclature
•
•
•
•
•
•
•
•
•
Kingdom Prokaryotae
Division
Gracilicutes
Class
Scotobacteria
Subclass
Order
Spirochaetales
Family
Spirochaetaceae
Tribe
Genus
Borrelia
Species
Borrelia burgdorferi
BACTERIA
Prokaryotic Cell Morphology
BACTERIAL CELL
• 50% protein
• 20% nucleic acids (10x more RNA than
DNA)
• 10% polysaccharides
• 10% lipids
Bacterial Chromosomes
• Single, circular, double-stranded DNA
(exception - borrelia = linear)
ori
C
ori
C
• Replication begins at unique point; bidirectional
• Haploid (1 to 4 copies depending on growth rate)
• 600 to >5000 kb* in size (smaller = more
dependent on host/environment)
• Up to 1 mm in length; supercoiled
• Contained in nucleoid
* ~1 kb/gene
Bacterial Nucleoids
• Chromosomal DNA (60%; 2-3% dry wt of cell) + RNA
(30%) + Protein (10%)
• No nuclear membrane
• No histones (~6 chromosome-associated basic proteins
involved in determining chromosomal structure)
• Polyamines (e.g., spermidine and putrescine) neutralize
negative charges on phosphates
• Haploid chromosome in cytoplasm
– 1 to 4 nuclear bodies/cell, number depends on growth rate
(faster = more)
• Can be membrane-associated (during cell division)
Bacillus cereus
Light Microscopy 2500x
Feulgen strain
Jawetz Med Micro 25e
Escherichia coli
Electron microscopy
Extrachromosomal DNA
• Plasmids - Replicate in cytoplasm, independent of
chromosome.
–
–
–
–
Double-stranded DNA; usually circular (borrelia = linear)
Few to several hundred kb
Few to several hundred copies per cell
Conjugative (F, R), antibiotic resistance,
metabolic, virulence
• Bacteriophage - virus;
– replicates in cytoplasm or integrates
into chromosome
– can contribute to virulence
into
Bacterial Structure
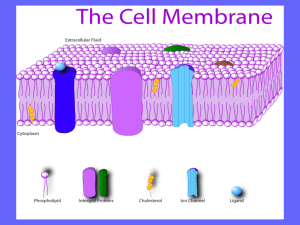
Cytoplasmic Membrane
• Lipid bilayer
–
–
–
–
–
Permeability barrier
Active transport
Electron transport
Oxidative phosphorylation
Photosynthesis
• Affected by antibacterials
– Detergents
– Polymyxins (damage PEcontaining membranes)
– Ionophores (disrupt
membrane potential)
Cell Wall
• Shape
• Barrier (osmotic
resistance)
• Comprised of highly
crosslinked peptidoglycan
• Affected by antibacterials
(e.g, b-lactam antibiotics,
lysozyme)
• Basis for gram-stain
Peptidoglycan
• Backbone of N-acetyl
glucosamine and Nacetyl muramic acid
• Cross-linked by
peptide bridges at
MurNAc
http://employees.csbsju.edu/hjakubowski/classes/ch331/cho/peptidoglycan.gif
Peptidoglycan
http://de.wikipedia.org/wiki/Peptidoglycan
Peptidoglycans
Transglycosylases
[GlcNAc-MurNAc]n
(TG) link
[GlcNAc-MurNAc]n
D-glu
D-glu
L-lys
L-lys
(gly)n
Amidases (autolysins, e.g.)
cleave
L-ala
L-ala
Hydrolases (lysosyme,
mutanolysin, e.g.) cleave
(gly)n
D-ala
D-ala Transpeptidases (TP) link.
b-lactams resemble TP substrates,
block crosslinking of growing chain
PG structures vary
between/among Gm+
and Gm-. This =
Gm+.
b-lactams and Peptidoglycan
Crosslinking
[GlcNAc-MurNAc]n
L-ala
D-glu
L-lys
D-ala
D-ala
non-crosslinked
peptidoglycan
R
Transpeptidase
NH
O C CH
HN CH3
HC CH3
HOOC
Terminal D-ala-D-ala
CH2
C O
NH
O C CH
N CH
S
C
HC
b-lactam
ring
HOOC (CH3)2
Benzylpenicillin
(penicillin G)
Gram Stain
• Gram’s crystal violet (CV)
• Potassium-iodide (KI)
• Ethanol - decreases hydration of cell wall
• Wash
CV-I complexes trapped in thick cell walls
(cells remain purple = gram-positive)
• Safranin (red)
thin cell walls don’t retain CV-I complexes,
counterstained with safranin
(red = gram-negative)
Exceptions to gram-positive /
gram-negative staining
• Mycoplasmas - no cell wall.
• Mycobacteria - lipid interferes with stain
– Detected with acid fast stain (carbol fuschin
retained following decolorization with
HCl/EtOH)
Both are related to gram-positives, based on
genetic analyses (rRNA sequence)
Gram-positives
•
•
•
•
•
Cytoplasmic Membrane
Cell wall
Lipoteichoic acid
Teichoic acid
Proteins
Gram-positive Cell Walls
• Thick peptidoglycan (10 to 100 nm)
• Wall teichoic acids (WTA) - repeating units of
phosphodiester-linked (negative charge) glycerol or
ribitol backbone + side chains (D-ala, glucose).
Covalently linked to PG (MurNAc)
Bacillus
subtilus
W23
TA Repeat
Ribitol-P
Linkage Unit (LU)
CH 2 OH
O-CH 2
H-C-O-R 1
H-C-OH
H-C-O-R 2
O
H-O-C-H
P
O
P
CH 2 O
O-CH 2
H-O-C-H
O-
OH
CH 2 OH
O
O
O
O-
O
CH 2 OH
O
O
P
CH 2 O
Peptidoglycan
HNAc
OH
HNAc
CH 2
P O
O
O
O
O - HO
GlcNAc-
HNAc
peptide
O(Glycerol-P)
CH 2 O
2 -( N-acetylmannosamine
)-GlcNAc-P-----MurNAc-GlcNAc--
(n)
**Glycerol-P
TAs also have linkage unit - R groups differ on
R1 = H or Ala; R2 = H or Glc
TA repeat and LU
Gram-positive Teichoic Acids
• Wall Teichoic Acids (WTA) – covalently linked to PG
• Lipoteichoic acids (LTA) – similar to WTA but anchored to
cytoplasmic membrane lipids; phosphodiester-linked (negative
charge)
• LTA and WTA
• ion binding
• charge maintenance
• membrane integrity
• adherence
• anchor proteins
• Cell walls - inflammation
Gram-negatives
• Cytoplasmic
membrane
• Cell Wall
• Outer membrane
• Lipopolysaccharide
• Proteins
Gram-negatives
• Cell Wall
– Thin peptidoglycan (1 layer; 2 nm)
– No WTA or LTA
• Periplasmic space - digestive and protective
enzymes; transport
• Outer membrane (OM) - blocks entry of large
molecules (>800 Da). Not typical lipid bilayer.
– Attached to PG by lipoprotein
– Lipopolysaccharide (LPS) - forms outer leaflet of OM
– OM proteins – transport; porins allow passive
diffusion of low MW hydrophilic compounds
(sugars, amino acids)
OmpF
Lipopolysaccharide (LPS)
• Endotoxin - toxic shock; fever. leukopenia, hypotension,
acidosis, DIC, death
(OM)-Lipid A --- core polysaccharide --- O Ag
toxic properties
HM HM
MM
LM
varies with species
polysaccharide
varies with strain
3 - 4 sugars/repeat
Up to 25 repeats
serotyping
Gram-negative Surface
(Cytoplasmic Membrane)
Optional Features (Gram +/-)
• Capsules - polysaccharide or protein (usually
covalently linked to peptidoglycan)
– Antiphagocytic (block C3b deposition or recognition),
attachment
• Surface Proteins - anchored in CM, OM, CW
– Antiphagocytic, attachment
• Flagella - protein. Rotates to propel cell.
Flagella - peitrichous
– Motility, chemotaxis, virulence (H-antigen)
capsules - colony
Flagella - EM
capsules - microscope
Flagella - unipolar
Optional Features (Gram +/-)
• Pili - protein. Shorter, narrower than flagella.
• Common - peritrichous; attachment
• F (sex) - single; gene transfer (conjugation; gram -)
• Toxins - excreted; act on host cells; Clostridium
botulinum; Vibrio cholerae
• Enzymes - hyaluronidase, proteases, DNases
• Endospores - dehydrated cells; Clostridium, Bacillus
species (gram +)
F-pilus