Force Field of Biological System
advertisement
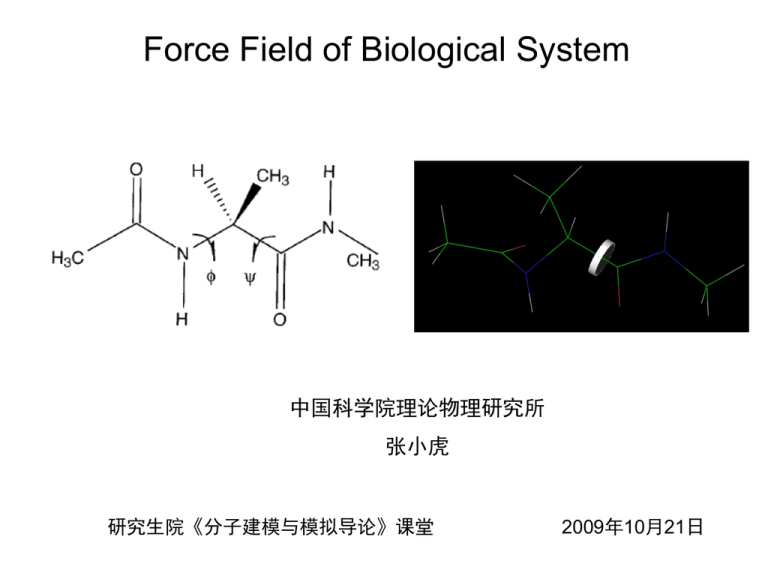
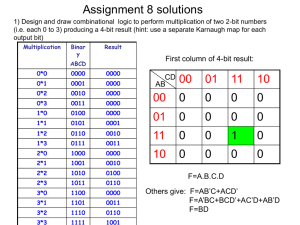
Force Field of Biological System 中国科学院理论物理研究所 张小虎 研究生院《分子建模与模拟导论》课堂 2009年10月21日 Why do we need force field? 1. Force Fields • • • • • Classical Newtonian Dynamics Electrons are in the ground state Force fields are approximate Nonbonded force fields for biological systems are effective pair potentials No Explicit term for hydrogen bonding References H. J. C. Berendsen, et al, Gromacs User Manual version 4.0 A. D. MacKerell, Jr. , et al, "Comparison of Protein Force Fields for Molecular Dynamics Simulations“ A. D. Mackerell, Jr. , et al, "Empirical Force Fields for Biological Macromolecules: Overview and Issues“ J. W. Ponder, et al, "FORCE FIELDS FOR PROTEIN SIMULATIONS“ Takao Yoda, et al, “Comparisons of force field for proteins by generalized-ensemble simulations” 2. Commonly used force fields • Amber: Assisted Model Building with Energy Refinement • CHARMM: Chemistry at HARvard Macromolecular Mechanics • OPLS-AA: Optimized Potentials for Liquid Simulations- All Atom • GROMOS: GROningen MOlecular Simulation References W. D. Cornell, et al (1995) ”A second generation force field for the simulation of proteins, nucleic acids, and organic molecules” A. D. MacKerell, et al (1998) ”All-atom empirical potential for molecular modeling and dynamics studies of proteins” W. L. Jorgensen, et al (1996) ” Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids” C. Oostenbrink, et al (2004) “A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6” 3. Functional forms Basic functionals Vtotal Vbonded Vnonbonded Vbonded K (b b ) b bonds Vnonbonded 0 2 K ( ) 0 2 angles R min,ij ij rij nonbonded pairs ij 12 Rmin,ij 2* rij K [1 cos(n )] dihedrals 6 qq i j rij 4. Differences for bonded interactions Valence Angles • • • • AMBER: k 2 CHARMM: + ( r r 0 ) 2 OPLS-AA: GROMOS: Improper Dihedral Angles maintain chirality or planarity Urey- Bradly angle term • • • • AMBER: + CHARMM: + OPLS-AA: + GROMOS: + k (1 cos 2w) k 2 / 2 k (1 cos 2w) k 2 / 2 5. Differences for nonbonded interactions Handling of 1,4-nonbonded interactions between A, D in dihedral A-B-C-D • • • • AMBER: CHARMM: OPLS-AA: GROMOS: LJ ½ Coulomb 1/1.2 not scaling except some special pairs LJ ½ Coulomb ½ case by case 6. How to construct a force field? Adjusting parameter values until the force field is able to reproduce a set of target data to within a prescribed threshold ; name bond_type mass charge ptype sigma epsilon amber99_0 H0 0.0000 0.0000 A 2.47135e-01 6.56888e-02 amber99_1 BR 0.0000 0.0000 A 0.00000e+00 0.00000e+00 amber99_2 C 0.0000 0.0000 A 3.39967e-01 3.59824e-01 amber99_3 CA 0.0000 0.0000 A 3.39967e-01 3.59824e-01 amber99_4 CB 0.0000 0.0000 A 3.39967e-01 3.59824e-01 amber99_5 CC 0.0000 0.0000 A 3.39967e-01 3.59824e-01 amber99_6 CK 0.0000 0.0000 A 3.39967e-01 3.59824e-01 amber99_7 CM 0.0000 0.0000 A 3.39967e-01 3.59824e-01 Target data Experimental: vibrational spectra; heats of vaporization; densities; solvation free energies; microwave, electron, or X-ray diffraction structure; and relative conformational energies and barrier heights. QM: vibrational spectra; minimum energy geometries; dipole moments; conformational energies and barrier heights; electrostatic potentials; and dimerization energies The Amber, CHARMM, GROMOS, and OPLS-AA force field for proteins each target a different subset of the possible experimental and QM data, although there is substantial overlap between the subsets. AMBER AMBER84: Polar hydrogens + united atoms ( hydrogens bonded to carbon) AMBER86: All- atom model • Based on experimental with gas phase simulation • Key ideas: ESP partial charge ( qi , qj ) ( Kb , b0 , Ksita , Sita0 ) from crystal structures, match NMF for peptide fragments VDW fits amide crystal data Dihedral match torsional barriers from experiments and quantum calculations AMBER94: Aimed to better perform Condensed phase simulations • Partial charges: Dependency on environments: RESP Dependency on conformations: fit simultaneously with multiple configurations • More accurate electron correlation method and larger basis set to determine torsional terms AMBER96,99 • Account long-range effects • Fit tetrapeptide + dipeptide AMBER03 • More accurate electron correlation method and larger basis set to determine torsional terms and partial charges • Continuum solvent models instead of vacuum CHARMM Key idea: Balancing water-protein, water-water, and protein-protein interaction energies in the condensed phase Difference: Dimerization OPLS-AA GROMOS energies, molecule-water minimum-energy distances 6. Comparison of force field in realization Favor Alpha-helix: Amber 94, 99 Beta-hairpin: GROMOS96 Intermediate: CHARMM22, AMBER96, OPLS-AA/L Experimental agreement Alpha-helix: • Remarkable agreement: Amber 99, CHARMM22 • Consistent with some experiments: AMBER96, OPLS-AA/L • Disagreement: AMBER94, GROMOS96 Beta-hairpin: • Remarkable agreement: OPLS-AA/L, GROMOS96 • Consistent with some experiments: AMBER96 • Disagreement: AMBER94, AMBER99, CHARMM22 THANK YOU