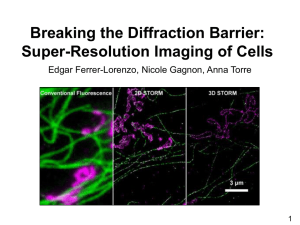
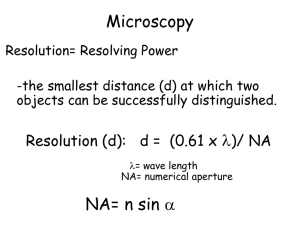
第一组-超分辨成像及应用
advertisement

超分辨成像及应用 深圳大学光电工程学院 第一小组:王广盛、吴茜茜、田蜜、袁淑怡 背景 光学显微镜的空间分辨率 背景 瑞利判据 实现超分辨成像的途径 利用倏逝波(近场) 增大数值孔径 利用荧光(双光子或多光子) 点扩展函数工程 分子定位 远场 概要 STED( Stimulated Emission Depletion fluorescence Microscopy ) (受激发射损耗显微技术) PALM( photoactivated localization microscopy ) (光激活定位显微) STORM( stochastic optical reconstruction microscopy) (随机光学重构显微技术) 探针 应用 概要 STED( Stimulated Emission Depletion fluorescence Microscopy ) (受激发射损耗显微技术) PALM( photoactivated localization microscopy ) (光激活定位显微) STORM( stochastic optical reconstruction microscopy) (随机光学重构显微技术) 探针 应用 STED( idea ) STED ( idea ) Stefan W.Hell 核心思想是利用受激福射选择性消耗激发光斑中边沿区域的激发态荧光分子 从而减少有效荧光的发光范围,压缩有效PSF尺度,提高系统分辨率; 点扩散函数 (Point Spread Function ,PSF ) 是一个无限小物点通过光学系 统在像平面处的光强分布函数; 原则上说 , 所有的光学超分辨技术都是围绕着如何得到一个尺度更小的 PSF STED (principle) Normal fluorescence occurs by exciting an electron from the ground state into an excited electronic state of a different fundamental energy level (S0 goes to S1) which, after relaxing back to the ground state (of s1), emits a photon by dropping from S1 to a vibrational energy level on S0. STED interrupts this process before the photon is released The excited electron is forced to relax into a higher vibration state than the fluorescence transition would enter, causing the photon to be released to be red-shifted as shown in the image Because the electron is going to a higher vibrational state, the energy difference of the two states is lower than the normal fluorescence difference. This lowering of energy raises the wavelength, and causes the photon to be shifted farther into the red end of the spectrum. STED (principle) 首先采用一束超短脉冲激光将爱里斑区域内的荧光分子激发至第一激发态的上 能级 S1’,电子很快通过振动弛豫至第一激发态的最低振动态S1; 接下来一束红移的圆环型STED光将爱里斑边沿处于激发态的荧光分子消激发至 基态的上能级S0’,振动弛豫回到S0。 爱因斯坦的受激发射辐射理论指出受激发射和受激吸收系数是相等的,STED光对 荧光分子产生的 S0’ → S1 受激吸收过程和 S1 → S0’受激发射过程是等概 率的。幸运的是振动弛豫速率为10 11 ~ 10 13 S-1, 荧光发射速率约为 10 9 S-1 , 换言之 ,S0’能级寿命比在S1短得多,处于S0’能级的电子迅速耗尽,避免了被消 激发回到S0’能级的电子的二次激发,因此S1 → S0’的受激发射过程占绝对的 优势。 爱里斑区域内仅没有被STED光照射的中心处的荧光分子可以正常发射荧光,PSF 尺度被大大压缩。 STED (principle) 一般来说, STED 显微术可实现的分辨率可由以下公式表示 d 2 NA 1 I max Is 其中,λ 为激发光的波长,NA为所用显微物镜的数值孔径,Imax为损耗光的最大光 强,Is为阈值光强。Is由所用样品的荧光寿命τ和损耗光的吸收截面σ决定具体为: Is =1/τ·σ 非常有趣的是虽然激发光和 STED光都是衍射受限的,但分辨率却与波长无关,只要 光强允许无限增加 , 分辨率没有极限。 STED(STED 光产生) STED (系统光路) STED (激光类型选择) 激光器有脉冲光和连续光两种类型 在STED显微术刚被提出的时候,所有的 STED系统都是基于脉冲激光来搭建 的 采用脉冲光源使得激发光和损耗光在时域上具有可分离性,使得受激发 射损耗的消光过程更便于操控; 由于STED的显微术的分辨率随着所用损耗光光强的增加而提高,在相同 的平均功率下,脉冲光具有比连续光更高的峰值光强。 弊:为了实现较好的消光效果,激发光和损耗光的脉冲宽度需要根据所用荧光样 品的不同而做相应的优化调整,一般典型值为激发光脉宽 < 80 ps,损耗光脉 宽约为 250ps,同时相应的激发光脉冲和损耗光脉冲之间也应具有一定的时间延 迟。因此,脉冲光 STED 系统中往往需要放置对脉冲进行展宽和同步的光学器件, 从而使得STED 系统变得非常的复杂和昂贵。 STED (激光类型选择) S. W. Hell 等人发现,当损耗光的消光速率远大于激发光的激发速率时 , 损耗光和激发光在时域上的可分离性将变得不再重要, 由此提出了第 一个采用连续光源作为损耗光的 STED 系统。 虽然连续光STED 系统所需的平均功率较大,但是其峰值光强相比脉冲光来 说要小很多,因此可以降低对荧光粉产生不必要的多光子激发的可能性。 同时,连续光STED 系统还可以实现比脉冲型 STED 更快的扫描速度。当采 用连续光作为损耗光时, 激发光可以是脉冲型,也可以是连续型, 从而 大大扩展了 STED 显微系统的光源类型选择范围。 STED (波长选择) 在一个典型的 STED 显微系 统中, 激发光和损耗光波长 的选择需要满足以下原则:激 发波长应选在所用荧光粉激 发谱的峰值波长附近, 以保 证较好的吸收;损耗光波长应 选在所用荧光粉发射谱的长 波拖尾处, 以避免损耗光对 样品的二次激发。然而在这 种波长选择方法下, 损耗光 波长处的受激发射截面 σ 较小, 使得相应的阈值光强 Is较大, 从而导致了所需的 损耗光强很高, 对样品的漂 白较为严重。 d I 2 NA 1 max Is 3D-STED HeLa细胞中Alexa 568标记的组蛋白H3:z堆栈(91层) 的惠更斯反卷积之后,进行表面3D重建,颜色相同的结 构定位在同一z-平面上。 http://www.leicamicrosystems.com/cn/ 3D-STED (4Pi-STED) STED (分辨率的提高) 尽可能地提高样品的抗漂白性,从而提高可用于样品成像的损耗光强。 2009 年,S .W. Hell等人利用氮缺位(NV)样品(不会漂白)实现了 5.8 nm 的横向分辨率。 通过提高显微物镜的数值孔径来实现。 2012 年S. W.Hell 小组利用固态浸没技术(NA =2.2)实现了2.4nm 的 横向分辨率 ,这也是至今有报道的 STED 显微可实现的最高分辨率。 G- STED(Gated- STED) 荧光寿命在STEDCW显微镜的 有效焦点中的分布。长寿命 态(红色)位于中心处,而 短寿命态(蓝色)位于外围。 时间门探测技术(Time gated detection) STED(成像深度的突破) 对较厚的样品进行成像时,照明光束的穿透深度将受到样品内部散射的影响, 同时由于光束在厚样品中传播时会产生较大的像差,使得生成的损耗光斑质量 很差,严重影响系统的分辨能力。 第一种方法是采用双光子激发来代替常规 STED 系统中的单光子激发方式 。 双光子激发方式中所用的激发光一般为近红外光, 穿透能力较强; 可以用相同的波长来实现双光子激发和受激发射损耗, 从而使得系统中只 需要一个激光器, 简化了系统 。 2012 年,意大利的 Diaspro 小组首次在实验中证明了单波长双光子STED 的可行性,实现了85nm的空间分辨率。 另一种提高 STED 成像深度的方法则是基于自适应光学的原理。 参考文献 Hell, S. W., J. Wichmann (1994): "Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy" Opt. Lett. 19, 780-782 We propose a new type of scanning fluorescence microscope capable of resolving 35 nm in the far field. We overcome the diffraction resolution limit by employing stimulated emission to inhibit the fluorescence process in the outer regions of the excitation point-spread function. In contrast to near-field scanning optical microscopy, this method can produce three-dimensional images of translucent specimens. Hell, S. W., S. Lindek, E. H. K. Stelzer (1994): "Enhancing the axial resolution in far-field light microscopy: two-photon excitation 4Pi-confocal fluorescence microscopy" J. Mod Opt. 41, 675-681. Hell, S. W., S. Lindek, C. Cremer, E. H. K. Stelzer (1994): “Confocal microscopy with an increased detection aperture: type-B 4Pi confocal microscopy” Opt. Lett. 19, 222-224. Klar, T. A., S. W. Hell (1999): "Subdiffraction resolution in far-field fluorescence microscopy" Opt. Lett. 24, 954-956. Honigmann, A., V. Mueller, H. Ta, A. Schoenle, E. Sezgin, S. W. Hell, C. Eggeling: "Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells" Nature Commun. 5, 1 – 12. Hänninen, P. E. and S. W. Hell (1994). "Femtosecond pulse broadening in the focal region of a two-photon fluorescence microscope." Bioimaging 2: 117-122. Klar, T. A., E. Engel and S. W. Hell (2001). "Breaking Abbe's diffraction resolution limit in fluorescence microscopy with stimulated emission depletion beams of various shapes." Phys. Rev. E 64: 066613, 066611066619. http://www.leicamicrosystems.com/cn/. 李 帅等.受激发射损耗显微术(STED)的机理及进展研究.激 光 生 物 学 报,第 22 卷第 2 期.2013. 郝 翔.受激发射损耗显微技术中 0 /π圆形相位板参数优化.光学学报,第 31 卷第 3 期.2011. 概要 STED( Stimulated Emission Depletion fluorescence Microscopy ) (受激发射损耗显微技术) PALM( photoactivated localization microscopy ) (光激活定位显微) STORM( stochastic optical reconstruction microscopy) (随机光学重构显微技术) 探针 应用 光激活定位显微技术(PALM) 2002 年,Patterson和Lippincott-Schwartz首次利用一种绿色荧光蛋白 (GFP)的变种(PA-GFP)来观察特定蛋白质在细胞内的运动轨迹。 随后,德国科学家Eric Betzig敏锐地认识到,应用单分子荧 光成像的定位精度,结合这种荧光蛋白的发光特性,可以来突 破光学分辨率的极限。 2006年9月,Betzig和Lippincott-Schwartz等首次在 Science上提出了光 激活定位显微(photoactivated localization microscopy, PALM)的概念。 2007年,Betzig的研究小组更进一步将PALM技术应用在记录两种蛋白质的相 对位置,并于次年开发出可应用于活细胞上的PALM成像技术来记录细胞黏附 蛋白的动力学过程。 光激活定位显微技术(PALM) 原理:PA-GFP来标记蛋白质 405nm激光激活(低能量) 488nm激光照射,单分子定位 488nm激光长时间照射以漂白 重复使用405nm和488nm激活和漂白,循环上百次 图像重构,获得超分辨图像 光激活定位显微技术(PALM) A B C D A,B,C,D,E,F展示了 实际实验过程及得到的原始 数据; A',B',C',D',E',F'展示 了用高斯拟合确定荧光分子 的中心,叠加产生了超分辨 率图像; (A, A')未激活任何荧光分子 时; (B,B')405 nm 的激光激活了4 个荧光子; (C, C')用488 nm 的激光观察 一段时间后漂白了第一轮激 活的4个荧光分子; (D, D')第二轮重新激活的另 外的几个荧光分子; (E, E')两轮激活的荧光分子组 成的图像; (F, F') E图的局部放大。 光激活定位显微技术(PALM) 分辨率:仅受限于单分子成像的定位精度,理论上可达1nm 量级 优点:可用于活细胞测量 缺陷:只能用来观察外源表达的蛋白质,而对于分辨细胞内 源蛋白质的定位无能为力 概要 STED( Stimulated Emission Depletion fluorescence Microscopy ) (受激发射损耗显微技术) PALM( photoactivated localization microscopy ) (光激活定位显微) STORM( stochastic optical reconstruction microscopy) (随机光学重构显微技术) 探针 应用 随机光学重构显微技术(STORM ) 2006 年底,美国霍华德- 休斯研究所的 华裔科学家庄晓薇实验组开发出来一种 类似于PALM的方法,可以用来研究细 胞内源蛋白的超分辨率定位,即随机光 学重构显微技术(stochastic optical reconstruction microscopy,STORM)。 Michael JF, et al., Nature Methods 3(10), 793 (2006) 随机光学重构显微技术(STORM) 随机光学重构显微技术(STORM) 2μm 2μm 2μm 2μm 随机光学重构显微技术(STOR) 分辨率(横向):7 nm 优点:可用来研究细胞内源蛋白的超分辨率定位 缺点:不能用于活细胞测量; 由于用抗体来标记内源蛋白并非一对一关 系,所以STORM不能量化胞内蛋白质分 子的数量 3D-STORM 3D-STORM 3D-STORM 分辨率(纵向):50nm 优点:简单 缺点:成像的穿透深度较浅,只能成像大约600nm深 度的生物样品; 生物样本与显微镜物镜之间折射系数的差别会 引入球差效应,在成像深度越深时z 轴估计的 偏差越大。 Summary 存在的问题: 分辨率有待提高; 所需时间较长,只能研究固定的或时间分辨率要求不高 的样品。 解决方案: 开发出量子效率更高、荧光更稳定、颜色不同的荧光蛋 白或荧光染料(提高分辨率) 开发新型的、高通亮的光可控开关荧光分子(接收到足 够多的光子) 采用时分复用技术(提高时间分辨率) 参考文献 1.S. Jia, J. Vaughan, X. Zhuang, "Isotropic three-dimensional super-resolution imaging with a self-bending point spread function", Nature Photonics 8, 302-306 (2014) 2.K. Xu, G. Zhong, X. Zhuang, "Actin, spectrin and associated proteins form a periodic cytoskeleton structure in axons", Science 339, 452-456 (2013) 3.I. Kim, W. Pan, S. Jones, Y. Zhang, X. Zhuang, D. Wu, "Clathrin and AP2 are required for Ptdlns(4,5)P2-mediated formation of LRP6 signalosomes", JCB 200, 419-428 (2013) 4.S-H. Shim, C. Xia, G. Zhong, H.P. Babcock, J.C. Vaughan, B. Huang, X. Wang, C. Xu, G-Q. Bi, X. Zhuang "Super-resolution Fluorescence Imaging of Organelles in Live Cells with Photoswitchable Membrane Probes", Proc. Natl. Acad. Sci. 109, 13978-13983 (2012) 5.H. P. Babcock, Y. Sigal, X. Zhuang, "A High-Density 3D Localization Algorithm for Stochastic Optical Reconstruction Microscopy", Optical Nanoscopy 1, 6 (2012) 6.吕志坚, 陆敬泽, 吴雅琼,陈良怡,《几种超分辨率荧光显微技术的原理和 近期进展》,生物化学与生物物理进展,2009, 36(12): 1626~1634 概要 STED( Stimulated Emission Depletion fluorescence Microscopy ) (受激发射损耗显微技术) PALM( photoactivated localization microscopy ) (光激活定位显微) STORM( stochastic optical reconstruction microscopy) (随机光学重构显微技术) 探针 应用 荧光探针 荧光探针的发光基础 (F)PALM/STORM所用荧光探针的基本要求 (F)PALM/STORM中使用的荧光蛋白 (F)PALM/STORM中使用的荧光染料 新型光开关荧光蛋白mGeos 单体光转化荧光蛋白mEos3s 荧光标记 小分子有机染料 无机染料 荧光标记技术 荧光半导体量子点 发光稀土元素 荧光蛋白 荧光的基本原理 参数 吸收光谱absorptiom spectrum 消光系数molar extinction coefficient 激发光谱extinction spectrum 发射光谱emission 量子产率quantum yield 斯托克斯位移Stokes Shift/反斯托克斯位移antiStokes Shift 这些参数决定一个荧光分子可不可以被用作荧光探针以 及如何选择激发和观察条件。 (F)PALM/STORM所用荧光探针基本要求 对于STED技术和SIM技术,一些常规的荧光蛋白和EGFP和 mCherry等都能很好的被应用。但是对于(F)PALM/STORM成像实 验,只有特殊的荧光探针才能很好的被应用。 1.荧光探针和标记蛋白最后一一对应 ; (对单分子定量研究尤为重要) 2.单分子具有极高的光量子产出和极低的背景荧光; (提高单分子的定位精度) 3.较低的标记密度; (以便于单分子定位) (F)PALM/STORM所用荧光探针基本要求 与荧光染料和量子点相比,荧光蛋白的最大优势就是一对一的标记 能力,荧光蛋白的数量与目的蛋白几乎一致,不存在错误标记的情 况。 荧光染料,通常具有十分优秀的光量子产出,很好的信噪比和 光稳定性。荧光染料的定位精度和成像效果是目前最好的。 (F)PALM/STORM中使用的荧光蛋白 到目前为止,已经有超过30种荧光蛋白被发现或者改造出 来进行超分辨成像,按照光学特性大致可以分为三大类: 1.光激活荧光蛋白PAFP; (在起始状态下处于按状态,荧光发射能力很弱;在紫外光 或者紫色光照射下会转变为亮状态,发射出明亮的荧光) 2.光开关荧光蛋白PSTP/可逆光激活荧光蛋白; (反复多次在暗状态和亮状态之间转化) 3.光转化荧光蛋白PCFP; (最常用到的荧光蛋白,在特定激活光照射下激发谱和发射 谱会从一个波长转化到另一个波长) 光激活荧光蛋白PAFP 光激活荧光蛋白PAFP 最早的光激活荧光蛋白PAGFP是在GFP基础上引入T203H位点获得 的,它是报道的第一个光谱特性受激光调控的荧光蛋白。 • • • PAGFP 优点: 在400nm激光照射下,488nm激发的绿色荧光会有约100倍左右的提升。 缺点: 在(F)PALM/STORM成像中,PAGFP的背景荧光很高,单分子光量子产出比 较低,标记密度也比较低,这些因素限制了它的应用。 但是对于绿色荧光蛋白而言,PAGFP是目前唯一的选择。 • 在光激活红色荧光蛋白中,PA-mCherry1和PA-TagRFP是最好的选择,这两个光 激活荧光蛋白的单分子特性都很好。 PA-mCherry1来自于mCherry的定点随机突变筛选,最大激发和最大发射分别是 564nm和595nm PA-TagRFP来自于TagRFP,荧光亮度相当于PA-mCherry1的三倍,亮状态和暗状 态的比值是540:1,是目前最好的做单分子轨迹的荧光蛋白。 PAFP光学特性 光开关荧光蛋白PSTP 光开关荧光蛋白PSTP是一个荧光蛋白大家族: 来自四聚体蛋白22G的单体Dronpa是这个家族中被研究最多的一 个荧光蛋白。 ① 当很强的490nm激光照射到Dronpa上时,它迅速转化到类似PAGFP 起始的状态 ② 当暗状态的Dronpa暴露在405nm激光条件下时,它又可以返回到 亮状态,这两种状态可以反复激活和关闭多次。 Dronpa的优缺点 在完全激活状态的荧光 亮度相当于EGFP的 2.5倍 单分子性不好,光量子 产出低,不适合做基于 单分子的超分辨成像 新型光开关荧光蛋白mGeos 对mEos2的His62 位的突变获得的 Dronpa标记的蛋白 很难的到高品质的 超分辨图像,其单 分子荧光太弱,有 时很难被图像处理 软件识别 荧光蛋白改造技术, 来获得一直单分子特 性更好的绿色PSTP 【mGeos家族研发】 ① mGeos-M和 mGeos-S的单分 子性质都非常好 ② PH敏感的mGeos-E ③ 反复开关多次的mGeos-ES mGeos-M 通过实验统计发现 mGeos-M在所有的 PSTP中拥有最高的 光子产出 在HeLa细胞 中标记微丝蛋 白beta-actin 光子数统计 光开关荧光蛋白PSTP EYFP也属于光开关荧光蛋白家族,EYFP的优缺点: 拥有良好的单分子定位 精度,首次被用到活体 细胞成像中。 反复激活能力很差,不 如Dronpa和mGeos家族 青色荧光蛋白mFTP0.7,是mFTP1.0的副产品: 可以反复激活关闭很多次 单分子特性并没有被很好的研究 红色光开关荧光蛋白: KFP1、mApple、rsCherry 、rsCherryRev、rsTagRFP等 其中mApple的单分子性质很好,适合做(F)PALM/STORM成像。 PSTP光学特性 光转化荧光蛋白PCFP 在特定激活光照射下激发谱和发射谱会从一个波长转化到另一种波长 光转化机制目前报道的主要有两种:光诱导剪切断键;光诱导氧化。 EsoFP是最早被用到(F)PALM/STORM成像中的荧光蛋白之一 ,它来自于珊瑚中 人们首先开发出第一代 单体EosFP, 即m EosFP, 但是它只能在30度以下 的温度条件下才能成熟, 无法转染到真核细胞中。 紧接着一系列EosFP单体 (mEos2\mEosFPthermo\ mEos3.1\mEos3.1\mEos3.2) 被开发出来 目前mEos3.2是被开 发出来最好的绿变红 光转化荧光蛋白 统计表 PCTP光学特性 PCTP光学特性 荧光染料 I. 荧光染料的单分子光量子产出都比较高,一般在 1000光子以上,甚至上万光子产出;与之相比荧 光蛋白的光量子产出通常在500左右; II.如何标记 抗体标记:一抗去识别目的蛋白 二抗标记有荧光染料去识别一抗 优点:抗体标记特异性好 缺点:抗体太大,本身就会导致10nm作用的误差 参考文献 1. 王成, 马俊领, 魏勋斌. 远场超分辨随机光重建显微镜(STORM)研究进展, 2. 3. 4. 5. 6. 光学技术,2011,37,1. 常浩. 新型超分辨荧光探针和定位算法的开发. 2013年中国科学技术大学博士 学位论文. Graham T Dempsey, Joshua C Vaughan. Kok Hao Chen, Mark Bates, Xiaowei Zhuang. Evaluation of florophores for optimal performance in localization-based super-resolution imaging. 2011, Nature Methods, 8: 1027–1036. 吕志坚, 陆敬泽, 吴雅琼, 陈良怡. 几种超分辨率荧光显微技术的原理和近期进 展. 生物化学与生物物理进展, 2009, 36(12): 1626-1634. Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010, 190(2): 165-175. Bates M, Huang B, Zhuang X. Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr Opin Chem Biol. 2008, 12 (5): 505-514. 概要 STED( Stimulated Emission Depletion fluorescence Microscopy ) (受激发射损耗显微技术) PALM( photoactivated localization microscopy ) (光激活定位显微) STORM( stochastic optical reconstruction microscopy) (随机光学重构显微技术) 探针 应用 Content 静态成像 动态成像 载体研究 脑成像等 其它 光刻蚀、生物探针等 静态成像 体外生物分子结构 如DNA、DNA蛋白等 细胞结构 如各类的细胞骨架等 细胞器 如内质网、溶酶体、细胞内外小泡以及线粒体等 质膜蛋白或膜相关蛋白复合物 如神经细胞轴突、突触结合蛋白集群等 动态成像 · 囊泡运输 · 细胞膜脂筏 · 等等 Mitochondria (线粒体) Figure 1. Simulations. (a) Point spread functions (PSFs) drawn to scale of (i) a confocal microscope (diffraction limited), (ii) a super-resolution (SR) microscope enabling a resolution of 30 nm in the optical plane and conventional resolution along the z-axis, and (iii) a super-resolution microscope enabling isotropic resolution of 30 nm. This resolution was chosen because it can be attained with various instruments and because antibody labeling using a primary and a dye labeled secondary antibody would increase any structure to about this size . (b–d) Simulations on how different labeled mitochondrial structures would appear in the microscopic image. To this end, the simulated data (top: 3D rendering of the data) were convolved with the respective PSFs. Shown are xy and xz sections. The simulated mitochondria have a diameter of 220 nm (b,c; inner membrane) and 280 nm (d; outer membrane). Scale bars: 500 nm. Figure 2. Super-resolution microscopy of protein distributions in mammalian mitochondria. (a) Two-color STORM images showing the interaction between mitochondria (magenta) and microtubules (green) .(b) Two-color STED microscopy of hVDAC3 and hexokinase-I [47]. (c,d) GSDIM and STED imaging of Mic60 (mitofilin) showing the peculiar ordered arrangement of the MICOS clusters . Scale bars: 1000 nm. Jakobs, S., C. A. Wurm: "Super-resolution microscopy in mitochondria" Curr. Opin. Chem. Biol. 20, 9 - 15 Neurobiology (神经生物学) Figure 1. STORM imaging of actin filaments in the axon. (A) Conventional wide-field fluorescence image of actin (green) and the dendritic marker MAP2 (magenta) in a fixed neuron. (B) Three-dimensional STORM image corresponding to the inset outlined by the yellow rectangle in (A), containing axons and devoid of dendrites. The periodic ring-like structure of actin wrapped around the circumference of the axon is clearly apparent. The white boxed insets display the y/z cross sections. Figure 2. Nanocluster organization of scaffolding proteins in postsynaptic areas containing AMPA receptors. The top row provides examples of PSDs resolved with PALM for mEos2-tagged PSD-95, GKAP, Shank3, and Homer1c. The color-coded representation depicts the local density of the proteins in PSDs, highlighting ‘hot spot’ areas. Scale bar, 200 nm. In most synapses, these scaffold molecules were enriched in one or two nanoclusters, as shown in the bottom left bar diagram displaying the relative frequency of PSDs with 0, 1, or more clusters. The bottom right figure displays the cumulative frequency distribution and the similar mean of cluster area for the different scaffold molecules. Willig, K. I., F. J. Barrantes: "Recent applications of superresolution microscopy in neurobiology" Curr. Opin. Chem. Biol. 20, 16 - 21 Neurobiology (神经生物学、动态) Figure 3. In vivo STED microscopy of F-actin along the dendritic shaft and in dendritic spines. The image is the maximum intensity projection of the raw data, and it corresponds to a dendrite in the molecular layer of the visual cortex recorded in a live mouse. The blue and green arrows point to two narrow parts in the spine. Right panels: Line profiles of the blue and green-labeled positions and their corresponding Lorentzian fits (FWHM, full-width half-maximum). Willig, K. I., F. J. Barrantes: "Recent applications of superresolution microscopy in neurobiology" Curr. Opin. Chem. Biol. 20, 16 - 21 STED+光镊 (DNA) Figure (a) Schematic of the measurement assay showing two optically trapped microspheres tethered by DNA. An excitation (EXC) beam (and superimposed STED beam) that is scanned over the DNA is shown in green. (b) Experimental setup showing beam paths and crucial components. For optical trapping (1,064 nm, dark red), two orthogonally polarized optical traps are independently steered using tip-tilt mirrors (M1 and M2), and microsphere displacements are measured on two position-sensitive detectors (PSDs) using back-focal-plane interferometry. The microsphere-to-microsphere distance is obtained from LED-illuminated complementary metal-oxide semiconductor (CMOS) camera images (875 nm, purple). For fluorescence, one laser system supplies STED (745-nm, violet) and excitation beams (640 nm, 543 nm and 467 nm, respectively red, green and blue, filtered from a supercontinuum spectrum (SC) using an acousto-optical tunable filter (AOTF)). These beams are fiber-coupled and delivered to the confocal tip-tilt piezo mirror scanner (M3) after being combined by dichroics (yellow). The descanned fluorescence signal (dashed lines) is collected on fiber-coupled avalanche photodiodes (APDs), and descanned excitation light can optionally be detected using a photomultiplier tube (PMT) by placing a pellicle beam splitter (BS, orange) in the common path. A STED stripe is formed by a binary phase plate (PP), and the excitation beams can optionally be circularly polarized using a λ/4 retarder (QWP). Black and red stars indicate planes conjugate to the objective and condenser back focal plane, respectively. CL, condenser lens; D1–D11, dichroic mirrors; OL, water-immersion objective lens; T1–T3, telescopes. Heller, I., G. Sitters,O. D. Broekmans, G. Farge, C. Menges, W. Wende, S. W. Hell, E. J. G. Peterman, G. J. L. Wuite: "STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA" Nature Meth. 10, 910 - 916 STED+光镊 (DNA) Figure (a) Experimental force-distance curve of λ DNA in PBS. The dashed curve shows the extendible wormlikechain model calculated using a persistence length of 55 nm, a contour length of 16.5 μm and a stretching modulus of 1,350 pN. DNA overstretching occurs near 65 pN. Inset, measured (symbols, s.d.) and calculated (curves) force and position noise (ΔF and Δx, respectively) as a function of measurement bandwidth (average trap stiffness = 0.57 pN/nm; F = 25 pN). Red, blue and gray, noise for singlebead detection; black, noise for differential detection; solid curves, 3.2-μm-diameter microspheres; dashed curves, 0.9-μm-diameter microspheres. (b) Confocal microscopy image of Sytox Orange–labeled λ DNA between two 3.2-μm microspheres (excitation (exc.), 543 nm). (c–e) Confocal microscopy images of individual EcoRV–Atto 647N (c; exc., 640 nm), Sytox Orange dyes (d; exc., 543 nm) and EGFP-labeled proteins (e; exc., 467 nm). (f) Simultaneous multicolor imaging of Sytox Orange, Sytox Blue and BsoBI–Atto 647N. (g,h) Confocal microscopy images of individual BsoBI–Atto 647N restriction enzymes bound specifically to optically stretched λ DNA in the absence (g) and presence (h) of 100 nM of free Atto 647N NHS-ester (the local concentration in the flow cell is estimated to be lower owing to adsorption, approximately 40 nM). Scale bars, 1 μm. The typical frame rate for a 25 μm × 2 μm field of view using 100-μs dwell time per 75-nm pixel is 1 Hz. Heller, I., G. Sitters,O. D. Broekmans, G. Farge, C. Menges, W. Wende, S. W. Hell, E. J. G. Peterman, G. J. L. Wuite: "STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA" Nature Meth. 10, 910 - 916 actin cytoskeleton(肌动蛋白微丝) Figure 1: Dual-objective 3D STORM resolves individual actin filaments in cells.(a) Dual-objective STORM image of actin (labeled with Alexa Fluor 647-phalloidin) in a COS-7 cell. The z positions are color coded (violet and red, positions closest to and farthest from substratum, respectively). (b) Close-up of boxed region in a. (c) STORM image of same area obtained by using only information collected by Objective 1 of dual-objective setup. Ke Xu, Hazen P Babcock & Xiaowei Zhuang : " Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton " Nature Methods 9,185-188 actin cytoskeleton(肌动蛋白微丝) Figure 2: Sheet-like cell protrusion comprises two layers of actin networks with distinct structures.(a) Dual-objective STORM image of actin in a BSC-1 cell. (g,h) Ventral and dorsal actin layers of cell in a. Ke Xu, Hazen P Babcock & Xiaowei Zhuang : " Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton " Nature Methods 9,185-188 Clathrin-coated pits (网格蛋白小窝) Figure 1: 3D STORM images of CCPs labeled with photoswitchable cyanine dyes via a SNAP tag in live cells. (a) Conventional wide-field image of CCPs in a live cell. (b) A 3D STORM image of the same area taken in 30 s. Only x-y projection is shown. (c) Conventional (left) and 3D STORM images of the boxed region in b. Cross-sections of CCPs at the indicated locations are shown on the right: x-y crosssection near the plasma membrane (top images) and x-z cross-section cutting through the middle of the CCP (bottom images). (d) A 3D STORM image taken in 1 s showing an x-y cross-section of two CCPs (left) and an x-z cross-section of the CCP indicated by the arrow. Scale bars, 1 μm (a,b) and 100 nm (c,d). Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang : “ Fast, three-dimensional super-resolution imaging of live cells” Nature Methods 8, 499–505 Clathrin-coated pits (网格蛋白小窝) Figure 2: Two-color 3D STORM images of CCPs and transferrin in live cells. (a) Conventional image of CCPs and transferrin in a live cell. CCPs were labeled with Alexa647 via a SNAP tag (magenta) and transferrin was directly labeled with Alexa568 (green). (b) A 3D STORM image x-y projection of the same area taken in 30 s. (c,d) STORM images of CCPs indicated in b: x-y cross-section near the plasma membrane (left), x-z cross-section cutting through the middle of the invaginating CCP (middle) and corresponding x-z cross-section of the clathrin channel only (right). Scale bars, 500 nm (a,b) and 100 nm (c,d). Sara A Jones, Sang-Hee Shim, Jiang He & Xiaowei Zhuang : “ Fast, three-dimensional super-resolution imaging of live cells” Nature Methods 8, 499–505 hippocampal neurons(海马神经元) Figure 1. Comparison between cytoplasmic and membrane labeling for neuron imaging. (A) STORM images of microtubules demonstrating the effect of label density. In the first panel the localizations from only the first few hundred frames of a STORM movie are included in the reconstructed image to simulate the effect that would be observed in the case of low label density. In the last panel localizations coming from the entire STORM acquisition are included to simulate the effect that would be observed in the case of high label density. The panels in between include progressively increasing number of localizations in the final reconstructed image. It is not possible to reconstruct the actual microtubule structure from the first image due to the low number of localizations, whereas the ability to reconstruct the microtubule structure increases with increasing number of localizations. (B) 2D STORM image of a neural process expressing YFP in the cytoplasm. The YFP was immuno-labeled with antibodies conjugated to photoswitchable A405-A647 pair for STORM imaging. The small volume of these processes results in a low localization density in STORM images. (C) 2D STORM image of a neural process expressing mCherry attached to the membrane through a palmitoylation sequence. The mCherry was similarly immuno-labeled。 The membrane targeting resulted in a 3.6-fold improvement in label density. Lakadamyali M, Babcock H, Bates M, Zhuang X, et al. : “3D Multicolor Super-Resolution Imaging Offers Improved Accuracy in Neuron Tracing.” PLoS ONE hippocampal neurons(海马神经元) Figure 4. Two-color and multicolor (Brainbow-like) imaging of hippocampal neurons by STORM. (A) A zoomed-in field of view of neural processes with (left) and without (right) the color information. The neurons were separately transfected with YFP and mCherry, mixed and co-cultured. (B) STORM image of neural processes labeled with a combination of three fluorescent proteins. The neurons were co-transfected with a mixture of the three fluorescent proteins. Each fluorescent protein was immuno-stained with antibodies conjugated to different dye pairs. The arrow and arrowhead point to two neural processes that show different color combinations. (C) The STORM image of the same region of neural processes (upper panels) is shown in the presence (left) and absence (right) of color. The tracing results for these two cases are shown in the bottom panels. (D) Tracing results with (left) and without (right) color information for the image shown in (B). Lakadamyali M, Babcock H, Bates M, Zhuang X, et al. : “3D Multicolor Super-Resolution Imaging Offers Improved Accuracy in Neuron Tracing.” PLoS ONE Escherichia coli chemotaxis network D.Greenfield,A.L.McEvoy,H.Shroff,G.E.CROOKS,: “Self-organization of the Escherichia coli chemotaxis network imaged.” PLoS ONE 参考文献 1. Jakobs, S, Wurm CA. Super-resolution microscopy in mitochondria. Curr. Opin. Chem. Biol. 2014, 20: 9 - 15. 2. Willig, K I, Barrantes FJ. Recent applications of superresolution microscopy in neurobiology. Curr. Opin. Chem. Biol., 2014, 20: 16 - 21. 3. Heller I, Sitters G, Broekmans OD, Farge G, Menges C, Wende W, Hell SW,Peterman EJ, Wuite GJ. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nature Meth. 2013, 10: 910 916. 4. Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nature Methods, 2012, 9:185-188. 5. Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011, 8(6): 499-508. 6. Lakadamyali M, Babcock H, Bates M, Zhuang X, Lichtman J. 3D multicolor superresolution imaging offers improved accuracy in neuron tracing. PLoS One.2012, 7 (1): e30826. 7. Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with superresolution light microscopy. PLoS Biol. 2009, 7(6): e1000137. 总 结 1、STED 成像技术的最大优点是可以快速地观察活细胞内 实时变化的过程,因此在生命科学中应用更加广泛.主要 缺陷在于光路复杂,设备昂贵,对系统的稳定性要求很高。 2.PALM 的成像方法只能用来观察外源表达的蛋白质,而 对于分辨细胞内源蛋白质的定位无能为力。 3、STORM方法可以用来研究细胞内源蛋白的超分辨率定 位。但是由于用抗体来标记内源蛋白并非一对一的关系, 所以STORM不能量化胞内蛋白质分子的数量,同时也不 能用于活细胞测量。 趋 势 1、当前的发展趋势是结合不同成像模式的优点,进一步提高 成像的精度和准确度。 2、另一个方面是继续开发出量子效率更高、荧光更稳定、颜 色不同的荧光蛋白或者是荧光染料。 3.通过改进荧光探针和成像模式来进一步提高活细胞上超分辨 率成像的时空分辨率。