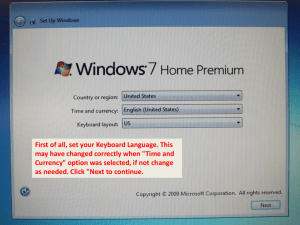
Project Overview
advertisement
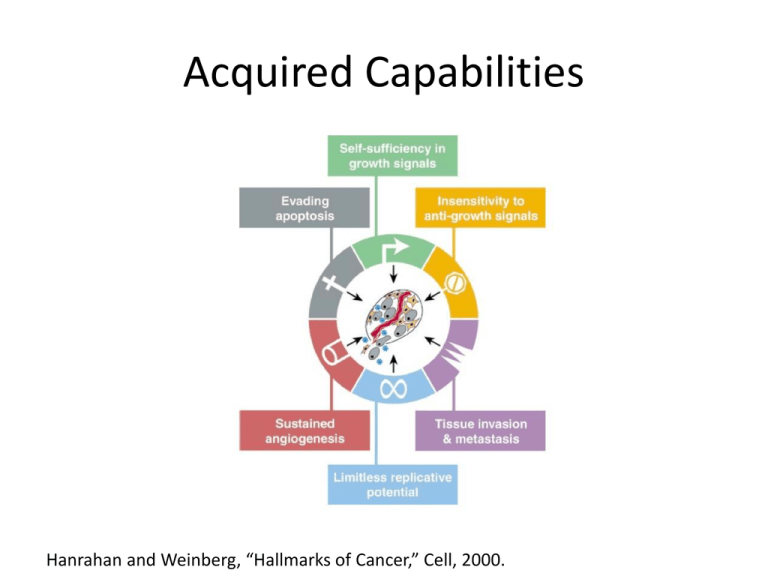
Acquired Capabilities Hanrahan and Weinberg, “Hallmarks of Cancer,” Cell, 2000. Pathways Hanrahan and Weinberg, “Hallmarks of Cancer,” Cell, 2000. Pathways • EGFR: DNA synthesis and cell proliferation – look for activation of Sos or Ras – Spratt, Balmick, Ratsimihah • TCR: antigen recognition – Look for activation of Erk – Packer, Rodriguez, Yalkabov • FcεRI: immune response – Look for activation of Syk – Feitzinger, Abbasi, Nunez • Apoptosis: cell death – Look for activation of – Miranda, Garib, Green • HMGB1: inflammation – Look for activation of ? – Weinberg, Zegel, Lagrange Project Goal • Develop a probability distribution for “first passage times” by running many simulations – First passage time is the time the target protein reaches a given level of activation • Variations: – – – – Vary ligand dose Vary level of activation Eliminate backward reactions Put some molecules in an “always-on” or “always-off” state • An experiment will consist of varying one thing and developing the probability distribution for each value Methodology • SSA simulations of assigned pathway using BioNetGen – Scripts developed for multiple runs and collecting data • Creates a directory for each run • Data collection program creates file for histogram – Use RuleBender for initial explorations – Find equilibrium values for inactive state with ODE – Find appropriate time interval (balance time to run with getting enough data) Methodology • Saving data – One directory for each experiment – One subdirectory for each value of variable – README in toplevel directory explaining what the variable and the values are plus any problems that arose – Histogram in top-level directory • In parallel: read about pathway – Wikipedia – Books – Papers Methodology • Splitting up the work • Other thoughts?? Pathways we won’t present T-Cell Receptors Alon, An Introduction to Systems Biology. Kinetic Proofreading in T-Cell Receptors Alon, An Introduction to Systems Biology. FceRI HMGB1








![Functional brain mapping of actual car-driving using [18F]FDG-PET](http://s3.studylib.net/store/data/008825166_1-520c765d189fcb1e600756a229ea56bc-300x300.png)