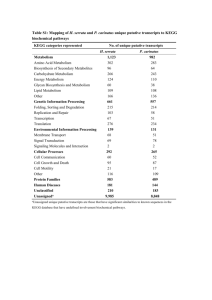
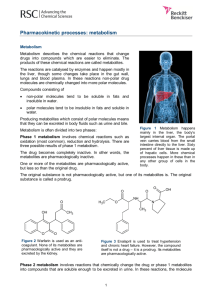
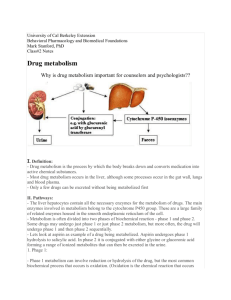
Three faces of metabolites: pathways, localizations and
advertisement

Three faces of metabolites: pathways, localizations and network positions Jing Zhao(赵静)1, Petter Holme2,3 1 Department of Mathematics, Logistical Engineering University, China 2 Department of Physics, Umeå University, Sweden 3 Department of Energy Science, Sungkyunkwan University, Korea The Fourth International Conference on Computational Systems Biology (ISB 2010) , September 9-11, 2010 Suzhou, China Outline Background Relation between the categories of metabolites Quantitative difference between categories Network structures of the categories Conclusion Metabolism metabolic network Subcellular compartments Cytoplasm, [c] Extracellular Space, [e] Mitochondrion, [m] Golgi Apparatus, [g] Endoplasmic Reticulum, [r] Lysosome, [l] Peroxisome, [x] Nucleus, [n] Pathways Carbohydrate Metabolism, C Energy Metabolism, E Lipid Metabolism, L Nucleotide Metabolism, N Amino Acid Metabolism, A Glycan Biosynthesis and Metabolism, G Metabolism of Cofactors and Vitamins, V Xenobiotics Biodegradation and Metabolism, X Biosynthesis of Secondary Metabolites, S Transport, T Modular topology of metabolic network Simulated annealing method r max M max [eii ( eij ) 2 ] i 1 j M: the modularity metric for a given decomposition of the network eij: the fraction of edges that leads between vertices of cluster i and j the sum is over the partition into clusters This study… Data used BiGG database Duarte, N. C.; Becker, S. A.; Jamshidi, N.; Thiele, I.; Mo, M. L.; Vo, T. D.; Srivas, R.; Palsson, B. O., Global reconstruction of the human metabolic network based on genomic and bibliomic data. PNAS 2007, 104, (6), 1777-1782. Relation between the categorizations M=0.676 Quantitative difference between categories: overlap score Prototypical overlap score XY (x, y) X (x) Y (y) x X y Y Normalized overlap score XY XY max( XX , YY ) X ,Y : two categorizations X(x) ,Y(y) : the fraction of metabolites in category x X, y Y, respectively XY(x,y): the joint frequency of x and y, i.e. the fraction of vertices that are categorized both as x X and y Y. Quantative difference between a feature of the real metabolic network and its randomized counterparts: Z-score f fr Z f r f : the metric of the feature in the real network fr : the mean of the corresponding metric in the randomized ensemble f r : the standard deviation of the corresponding metric in the randomized ensemble Overlap scores between categories Localization vs. network clusters : = 0.72, z =108.46; Network clusters vs. pathways: = 0.37, z = 45.94; Pathways vs. localization : = 0.42, z = 54.69 Network structure of the categories of the three categorizations Conclusions • We have investigated three ways of grouping metabolites according to pathways, localization, and network clusters respectively. • The compartmentalization is clearly organized into a core of extracellular media and cytoplasm, and a periphery of organelles; while the core and periphery modules of the pathway interactions correspond to housekeeping and advanced specific functions respectively. • Qualitative and quantitative analysis shows that the three categories are significantly correlated with each other, suggesting the interrelationship between biochemical specific functions, spatial isolations in cells, and topological compact regions in metabolic networks. Acknowledges National Natural Science Foundation of China (10971227) Swedish Foundation for Strategic Research Swedish Research Council the WCU program through NRF Korea funded by MEST R31–2008–000–10029–0 Thanks!