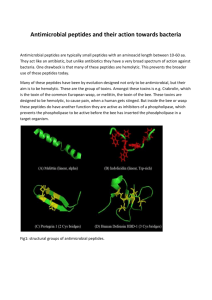
Database search introduction
advertisement

Database Searches Peptide mass fingerprinting digest Spectrum processing MS Protein (gel band or spot) Mass spectrum peptides Protein X theoretical digest 1554.25 2055.39 1942.44 1755.67 987.55 855.34 677.68 Peak list Search Protein Y theoretical digest Report HIT Protein X Protein Y Protein Z SCORE 1000 50 5 Protein Z theoretical digest Data processing …before searching • Baseline correct • Noise reduction • Peak deisotope for monoisotopic peaks …what is a monoisotopic peak • Copy to Excel and sort for peak intensity • Search with the top 75 peak ions (peptides) Generate mass list of all peptides • Computer program used to convert graph data (from mass spectrum) to table data (list of peptide masses) V o y a g e r S p e c # 1 [ B P = 8 4 2 . 5 , 2 2 2 0 7 ] 1045.6139 100 7 2 8 2 .1 972.5954 1007.467 973.4202 1023.462 973.4202 1036.481 975.6104 1066.493 977.4627 1082.488 982.4603 1105.601 983.4991 1153.524 998.4552 1172.581 1473.8500 90 80 2212.1514 % Intensity 70 1155.6985 60 50 1494.7859 40 1524.8172 1908.0468 1179.6516 30 1107.5970 20 1003.5450 10 1000.0361 0 1000 1666.8767 1310.7056 1608.9940 1324.7029 1788.0128 1477.8171 1210.6405 1718.8852 1215.0833 1425.9303 1600 2054.0554 2300.2157 2226.1625 2104.1794 2081.0745 2692.3379 2385.0063 2313.1521 2550.2102 2200 2788.4984 2800 M ass (m/z ) 3024.5809 3294.0001 3400 3603.6864 0 4000 Intensity Simple Mass Spectrum 771.9 1165.2 361.4 607.7 320.3 992.1 681.7 200 400 600 800 1000 Mass (Da) 1200 1400 1600 320.3 361.4 607.7 681.7 771.9 992.1 1165.2 List of masses Database search using computer algorithm e.g. MASCOT EAT EGR ISPYK EMETR EMANYK PLEASEMAK EATSTHEYAR Sequence matches PLEASEMAKEMANYKRISPYKREMETREATSTHEYAREGREAT Mass list of all peptides in the mass spectrum copied into here www.matrixscience.com Coverage = 26% We found peptides that make up only ¼ of the entire protein – but this is enough for a confident identification! QUESTIONS