Gene x Environment Interactions
Brad Verhulst
(With lots of help from slides written
by Hermine and Liz)
September 30, 2014
What does a GxE interaction in a twin
model really mean?
• Univariate Analysis: What are the contributions of A,
C/D & E to the variance?
• Heterogeneity Analysis: Are the contributions of
genetic and environmental factors equal across
different groups, such as sex, race, ethnicity, SES,
environmental exposure, etc.?
• Moderation Analysis: Are the contributions of genetic
and environmental factors to the variance constant
across the range of a second (moderator) variable?
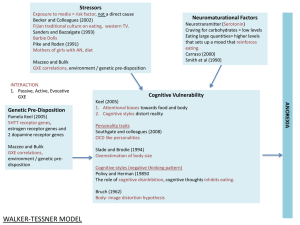
Gene-Environment Interaction
GxE
• genetic control of sensitivity to the environment
• environmental control of gene expression
– (environmental modulation of non-genetic paths)
Examples:
• Does heritability of IQ depend on SES?
• Does heritability of ADHD depend on age?
• Does the role parental monitoring depend on
genotype?
Gene-Environment Correlation
rGE
• genetic control of exposure to the environment
• environmental control of gene frequency
Examples:
• Active rGE: Children with high IQ read more books
• Passive rGE: High IQ parents give their children books
• Reactive/Evocative rGE: Children with ADHD are
treated differently by their parents
Moderating Variables
• Almost any variable can be used as a moderator…
… but be careful as not all variables make sense
as moderators (or are easy to interpret)
• If a variable has a genetic component (A > 0)
interpreting the GxE path is complicated by the
fact that the moderator is a function of both G &
E.
• Is it a GxG or a GxE interaction?
Heterogeneity Moderation
• An easy (but much less powerful) method of conducting GxE
• For categorical variables, estimate separate parameters for each
group.
– Sex Limitation is a classic case of GxE where separate parameters are
estimated for each group
– This can be extended to any number of categories (but quickly gets
tedious and difficult to interpret)
• This approach would not work for continuous variables (as there
are no discrete categories)
– Age
– Factor Scores of X, Y & Z
• Grouping these variables into categories loses a lot of information
and power
GxE Model & Theory
Purcell 2002 Twin Research
GxE Application
Turkheimer et al. 2003 Psychological Science
Turkheimer et al. 2003 Psychological Science
Definition Variables in OpenMx
• General definition: Definition variables are variables that
may vary per subject/pair and are not dependent variables
• In OpenMx: Specific values of definition variables for a
specific individual/pair is read into mxMatrix when
analyzing data of that particular individual/pair
Common Use of Definition Variables
• To model main effects of on the means (e.g. age and sex)
• To model changes in variance components as function of
some moderator variable (e.g. age, SES)
Cautionary Note about Definition
Variables
• Definition variables should not be missing if
dependent variable is not missing
• Definition variables should not have the same
missing values as dependent variable (e.g. use 2.00 for definition variable and -1.00 for
dependent variable)
• It is helpful to have very large values for missing
definition variables (so that if things go wrong the
results are unmistakably funky)
Definition Variables as Main Effects
General model with age and sex as main effects:
yi = α + β1(Agei) + β2(Sexi) + εi
Where:
yi is the observed score of individial i
α is the intercept or grand mean
β1 is the regression weight of age
Agei is the age of individual i
β2 is the deviation of females (if sex coded 0:males, 1:females)
Sexi is the sex of individual i
εi is the residual not explained by definition vars
(and can be decomposed further into ACE etc.)
Allowing for Main Effect
Means Vector
M + Xβ M + Xβ
Covariance Matrix
a2 + c 2 + e 2
H * a2 + c 2
H * a2 + c 2
a2 + c 2 + e 2
Allowing for Moderation
Means Vector
M + Xβ M + Xβ
Covariance Matrix
(a + Xϒa)2 + (c + Xϒc)2 +
H*
(e+ Xϒe)2
(a + Xϒa)2 + (c + Xϒc)2
H*
(a + Xϒa)2 + (c + Xϒc)2 +
(a + Xϒa)2 + (c + Xϒc)2
(e+ Xϒe)2
Existing Gene-Environment Interaction
Models
MZ=1
Classical Twin
Design
Purcell (2002)
DZ = ½
1
1
1
A
A
1
a + βaM
1
C
C
1
1
c + βcM
E
E
Basic Means
e+ β M
and Variances
e + β eM
Means
Moderation
Model
c + βcM
e
Pt1
Pt2
μ + Mβm
μ + Mβm
1
a + β aM
Example: Turkheimer Study
• Moderation of
unstandardized
variance
components
• Moderation of
standardized
variance
components
Cautions about interpreting the
Parameters
Unstandardized (UV) vs Standardized (SV)
Environment 1 Environment 2
Unstandardized
Variance
Standardized
Variance
Unstandardized
Variance
Standardized
Variance
Genetic
60
.60
60
.30
Common
Environment
35
.35
70
.35
Unique
Environment
5
.05
70
.35
Total Variance
100
1.00
200
1.00
Cautions about interpreting the
Parameters
Parameters are Conditional
• The estimated values of a, c & e in a Purcell model depend on the
value of the intercept (or the mean).
• If the mean is 0, the interpretation of the direct effect of a (or c) on
the phenotype is the genetic (or common environment) variance at
the mean.
• If the mean is not 0, the interpretation of the direct effect of a (or c)
on the phenotype is the genetic (or common environment) variance
is more complicated.
• Therefore, it is always suggested that the variance components are
plotted across the range of the moderator.
GxE in context of rGE
• If there is a correlation between moderator
(environment) and outcome, and you find a
GxE effect, it is not clear if:
– the environment is moderating the effects of
genes
OR
– trait-influencing genes are simply more likely to be
present in that environment
Ways to deal with rGE
• Limit study to moderators not correlated with
outcome
• Put moderator in means model to remove
covariance genetic effects shared by trait and
moderator
• Explicitly model rGE in bivariate framework