2.4
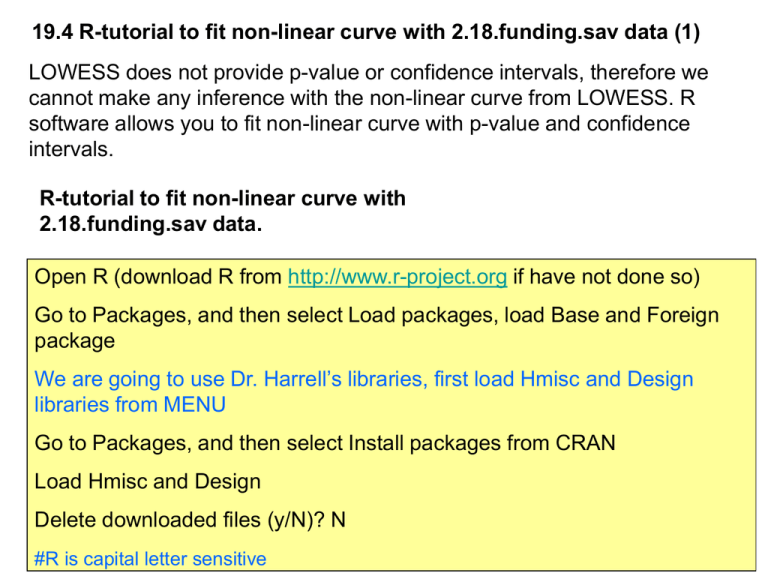
19.4 R-tutorial to fit non-linear curve with 2.18.funding.sav data (1)
LOWESS does not provide p-value or confidence intervals, therefore we
cannot make any inference with the non-linear curve from LOWESS. R
software allows you to fit non-linear curve with p-value and confidence
intervals.
R-tutorial to fit non-linear curve with
2.18.funding.sav data.
Open R (download R from http://www.r-project.org if have not done so)
Go to Packages, and then select Load packages, load Base and Foreign
package
We are going to use Dr. Harrell’s libraries, first load Hmisc and Design
libraries from MENU
Go to Packages, and then select Install packages from CRAN
Load Hmisc and Design
Delete downloaded files (y/N)? N
#R is capital letter sensitive
R-tutorial to fit non-linear curve with 2.18.funding.sav data (2)
# There is another step in order to complete loading job.
# at the command line “>” type the following
# (you can cut and paste the following commands from here to R):
library(Hmisc)
library(Design)
library(foreign)
library(MASS)
# See what are in there by typing
library(help="Hmisc")
library(help="Design")
# Now look in Hmisc for a command to read SPSS file
help.search("bootstrap") #general search
# R can be used as calculator
1+2
[1] 3
# a に1を代入
a<-1
# b に2を代入
b<-2
a+b
[1] 3
a*b
[1] 2
b ** b
[1] 4
R-tutorial to fit non-linear curve with 2.18.funding.sav data (3)
#If you want to know instruction how to use spss.get to read SPSS file
?spss.get
#Now let’s read in the dataset (first, you need to move the file to the
#directory called c:\\temp, if you want to use the following command,
#otherwise, specify the directory you stored the dataset.)
support<-spss.get('c://Rdata//support.sav', lowernames=T )
# List name of variables
names(support)
[1] "age"
"sex"
"hospdead" "slos" "d.time" "dzgroup"
[7] "dzclass" "num.co" "edu"
"income" "scoma" "charges"
[13] "totcst" "totmcst" "avtisst" "race" "meanbp" "hrt"
[19] "pafi" "bili" "crea" "ph"
"wblc" "resp"
[25] "temp" "alb"
"sod"
"glucose" "bun"
"urine"
[31] "adlp" "adls" "pre.1" "pred.cat" "rand.num" "rrand.nu"
[37] "filter.." "pre.2" "death" "years" "year3" "status3"
#if you want to know the contents of the datasets, type name of the dataset
support
age
1
2
3
4
5
6
7
8
9
10
11
12
13
sex hospdead slos d.time
43.53998 female
0 115
63.66299 female
1
14
41.52197 male
1
21
89.58795 male
1
4
67.49097 male
0
24
72.83795 male
1 109
75.36798 male
1
13
37.71899 male
1
7
58.95999 female
0
26
25.48700 male
1
19
56.66498 male
1
14
38.88300 male
0
15
66.54596 male
1
45
2022
14
21
4
1951
109
13
7
1882
19
14
1807
45
dzgroup
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
MOSF w/Malig
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
ARF/MOSF w/Sepsis
R-tutorial to fit non-linear curve with 2.18.funding.sav data (4):
R-output from describe function
#describe is tremendously useful to see what is in the dataset
describe(support)
42 Variables
1000 Observations
------------------------------------------------------------------------------------------------------------------age : Age
n missing unique Mean .05 .10 .25 .50 .75 .90 .95
1000
0 970 62.47 33.76 38.91 51.81 64.90 74.50 81.87 86.00
lowest : 18.04 18.41 19.76 20.30 20.31, highest: 95.51 96.02 96.71 100.13 101.85
------------------------------------------------------------------------------------------------------------------sex
n missing unique
1000
0
2
female (438, 44%), male (562, 56%)
------------------------------------------------------------------------------------------------------------------hospdead
n missing unique Sum Mean
1000
0
2 253 0.253
------------------------------------------------------------------------------------------------------------------slos : Days from study enrollment to hospital discharge
n missing unique Mean .05 .10 .25 .50 .75 .90 .95
1000
0
88 17.86
4
4
6
11
20
37
53
lowest : 3 4 5 6 7, highest: 145 164 202 236 241
-------------------------------------------------------------------------------------------------------------------
# To create nice graphs to describe data
datadensity(support)
#Scatter plot
2e+05
0e+00
totcst
4e+05
plot(totcst~alb, data=support)
1
2
3
4
5
#Scatter plot
9 10
8
7
log(totcst)
12
plot(log(totcst)~alb, data=support)
1
2
3
alb
4
5
# Save a new variable, ln.totcst into support data
support$ln.totcst <- log(support$totcst + 1)
#Type the next 2 lines before you do any graphical work
dd <- datadist(support)
options(datadist='dd')
# Lowess curve
plsmo(support$alb, support$ln.totcst, datadensity=T)
# Fitting linear regression
f.linear<-ols(ln.totcst~alb, data=support)
# Show results of the regression
f.linear
ols(formula = ln.totcst ~ alb, data = support)
Frequencies of Missing Values Due to Each Variable
ln.totcst
alb
105
378
n Model L.R.
565
66.88
Residuals:
Total RCC cost
Min
1Q
-9.93285 -0.77134
Median
0.02745
d.f.
1
R2
0.1116
3Q
0.74721
Max
3.07144
R2
Sigma
1.153
Coefficients:
Value Std. Error
t Pr(>|t|)
Intercept 11.1960
0.18971 59.017 0.000e+00
alb
-0.5263
0.06258 -8.411 4.441e-16
Residual standard error: 1.153 on 563 degrees of freedom
Adjusted R-Squared: 0.11
P-value
# Plot result of the linear regression
plot(f.linear, alb=NA)
# Check normality of residuals
hist(f.linear$residuals)
#Fitting non-lineaer linear regression
f.nonlinear<-ols(ln.totcst~rcs(alb,3), data=support)
# Graph the non-linear regression
plot(f.nonlinear, alb=NA)
#Viewing the result of the regression
anova(f.nonlinear)
Overall effect of ALB
p<0.0001 indicates
significant effect by
ALB
Analysis of Variance
Response: ln.totcst
Factor
d.f. Partial SS MS
F
alb
2
96.384725 48.192363 36.29
Nonlinear
1
2.322377 2.322377 1.75
REGRESSION
2
96.384725 48.192363 36.29
ERROR
562 746.263561 1.327871
P
<.0001
0.1865
<.0001
P<0.05 indicates
non-linearity
P<0.05 indicates
the model is
useful
Box-Cox transformation (1): Finding an optimal choice for power
transformation in R
A useful method so called Box-Cox transformation, will help you to identify the
optimal power transformation to achieve normality of residuals. Now we find
the best transformation for a regression of Crea= age
f.linear.crea<-ols(crea~age, data=support)
hist(f.linear.crea$residuals)
anova(f.linear.crea)
plot(f.linear.crea, age=NA)
Analysis of Variance Table
Response: crea
Df Sum Sq Mean Sq F value Pr(>F)
age
1
0.13
0.13
0.045 0.832
Residuals 995 2915.09
2.93
Box-Cox transformation (2): Finding an optimal choice for power
transformation in R
f.linear.crea<-lm(crea~age, data=support)
bcout<-boxcox(f.linear.crea)
bcout$x[bcout$y == max(bcout$y)]
[1] -0.5050505
Indicates that you may try
transfomation by Y-0.5
# Create a new variable
support$crea05<-support$crea**(-0.5050505)
Box-Cox transformation (3): Finding an optimal choice for power
transformation in R
# Re-do linear regression with transformed variable
f.new<-ols(crea05~age, data=support)
# Check residuals again
hist(f.new$residuals)
# Now you can see p-values
anova(f.new)
plot(f.new, age=NA)
Analysis of Variance
Factor
d.f. Partial SS
age
1
1.198304
REGRESSION
1
1.198304
ERROR
995 68.590890
Response: crea05
MS
F
P
1.19830356 17.38 <.0001
1.19830356 17.38 <.0001
0.06893557
Homework assignment 1
Using Support.sav and use R-software, answer the following
questions.
1. Plot non-linear regression slope of log-transformed total cost
by serum albumin level with 95% CI for the slope.
(a) Does R2 improve from the analysis of 19.1.1?
(b) Does the test of non-linearity for serum albumin level
suggest non-linear effect of serum albumin level?
(c) Is there association between transformed total cost and
serum albumin level?
Homework assignment 2
Using Support.sav and use R-software, answer the following questions.
2. Plot simple non-linear regression slope of log-transformed total cost
by SUPPORT coma score.
(a) Does R2 improve from the analysis of 19.2.1?
(b) Does the test of non-linearity for SUPPORT coma score
suggest non-linear effect of SUPPORT coma score?
(c) Is there association between transformed total cost and
SUPPORT coma score?