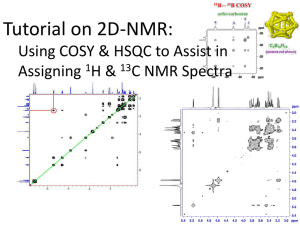
RAJ BHASKARAN, CLAFLIN UNIVERSITY “Establishing a NMR
advertisement

Establishing a NMR Structural Biology Center at Claflin University. Raj Bhaskaran Department of Chemistry Claflin University, Orangeburg, SC-29115 Clemson CI Symposium - Feb 13, 2013 NMR in CI Symposium Aim of this talk is two fold: To attract the Cyber Specialists to join hands with Claflin NMR team to mutually collaborate in the areas that require enormous computing and CI efforts; To attract Life Science researchers to interact with the NMR team to explore their problems structurally to provide a functional meaning; . . . to initiate GEAR CI collaborations. Clemson CI Symposium - Feb 13, 2013 Claflin’s NMR Route to Structural Biology and Drug Discovery Research Claflin has the high field 700 MHz NMR Spectrometer- BioNMR/ Metabolomics NSF Major Research Instrumentation grant is applied for a Cryoprobe and Sample Changer Solution Structure Determination and Protein Dynamics- Functional Interpretation based on structural investigations Protein (Enzyme) - Small Molecule (Substrate / Inhibitor) Interactions Protein Structures Derived Functional Interpretation on t. Thermophilus proteome, from Riken, Japan and Filariasis Nematode, Brugia Malayi proteome, (NIH, UIC) Rapid Protein Structure Determination for Structural Genomics Consortium: Structural Bioinformatics (North East Structural Genomics Consortium; Seattle Structural Genomics Center for Infectious Diseases) Computer Aided Drug Discovery: Virtual screening, Data Mining, QSAR Structure Based Drug Design: SAR by NMR, NMR Screening Clemson CI Symposium - Feb 13, 2013 Structure Based Drug Design •Suitable protein target New Compound •Structure of the target protein Biological Activity •Implementation of an easy and reliable HTS assay •Identification of a lead compound Suggests New Interaction Drug-Protein Complex •Computer assisted methods for estimating the affinity of new compounds •Access to a synthetic route to produce designed compounds. •Knowledge on shape of the pocket to design the shape of the drug. •The use of X-ray / NMR structures of Protein-Drug Complexes to design better-fitting, and hence more potent inhibitors Clemson CI Symposium - Feb 13, 2013 Ligands in the Drug Design Process Identified & Validated Target (genomics, transgenics etc) Protein Production Assays /HTS Structure Ligand-Protein structure Free state low affinity, high Kd compound libraries natural products directed libraries Hits more chemistry more structure Optimised Ligand high affinity, low Kd Clemson CI Symposium - Feb 13, 2013 Chemical tractability Toxicity DrugMetabolism / PharmacoKinetics Extensive computations involved in Protein NMR Study Isotopically 15N, 13C, Labeled NMR sample Acquisition of 3D heteronuclear (H, 15N, 13C) NMR experiments Backbone (HN, 15N, 13Ca, Ha ) and Side Chain (13Cb, Hb, 13Cg, Hg , …..) resonance assignment Unambiguous assignment of Nuclear Overhauser Enhancement ( proton-proton interaction within 5 A0 radius) Structure Determination using Restrained Molecular Dynamics and Simulated Annealing Protocol Structural Refinement using additional experimental restraints Protein Dynamics from 15N, 13C Relaxation experiments Clemson CI Symposium - Feb 13, 2013 NMR EXPTS SOFTWARE Backbone VNMR NHSQC; CHSQC; TOPSPIN TROSY HNCOCA; HNCA HACACONH; HNNCAHA NMRPIPE CBCACONH; HNCACB SPARKY HNCO; HCACOCANH AUTO ASSIGN Side chain PYMOL / MOLMOL HBCBCGCD(CE)HD(HE) SHIFTX CCH TOCSY 105 F 110 R E M P G G 115 P V W R K H Y I T 120 Y R I N N 125 Y T 130 P D M N R E D V D Y A 135 I R K 140 A F Q V W S 145 N V T P 150 L K F S K 155 I N T G M A 160 D I 165 L V V F 170 A R G A H G 175 D F H A F 180 D G K G G I daN(i, i+1) HCCCONH; CCONH dNN(i, i+1) HCCH COSY; daN(i, i+3) TALOS-PLUS Secondary Structure P G S G 195 I G G D A H 200 F D E D E 205 F W T 210 T H S G G T N 215 L F L T A V 220 H A I G H 225 S 230 L G L G H S S 235 240 245 D D K A V M F D T Y K Y V D I 250 N T F R L 255 S A D D 260 I R G I Q S L Y G daN(i, i+1) CYANA dNN(i, i+1) dab(i, i+3) XPLOR-NIH daN(i, i+3) NOE N15 EDITED 3D NOESY Secondary Structure MMP-12 Residue 0.6 HADDOCK 0.4 0.2 0.0 C13 EDITED 3D NOESY F G dab(i, i+3) daN(i, i+2) HCCH TOCSY (aro) A H A CSI daN(i, i+2) 190 HCCH TOCSY 185 L MODELFREE - 0.2 - 0.4 110 NoesyChsqc (aro) HUMAN ( E219A) MMP-12: Clemson CI Symposium - Feb 13, 2013 130 120 150 140 1H-15N 170 160 190 180 210 200 230 220 250 240 260 PROCHECK/PSVS HSQC SPECTRUM / NOE CONNECTIVITIES / CSI Bhaskaran & VanDoren JBNMR (2006) First NMR structure of the MMP-12 catalytic domain sans inhibitor RCSB PDB code 2POJ 2803 NOEs Used BB RMSD 0.32 A SC RMSD 0.68 A Ramachandran plot statistics: 77.3% in most favored regions 21.5% in allowed regions 0.0% in disallowed regions active site helix B & b-strands I, III are closer in free state NMR – Green; Inhibitors pull aB away from the far side X-RAY – Red; Clemson CI Symposium - Feb 13, 2013 29 unique NOEs ( dotted lines) account for the conformational adjustment Bhaskaran et al. J Mol Biol (2007) Protein Dynamics - rigidity of catalytic domain affect activity upon elastin & a1-AT. O rd er p a ra m eter S 2 O rd er p a ra met er S fo r M M P -1 2 b 1 .2 1 .1 b 1 a b2 b3 b4 a b5 2 fo r M M P-3 1 .2 a b b b b a a 1 .1 a 1 .0 1 .0 0 .9 0 .9 0 .8 0 .8 0 .7 2 0 .7 2 S 0 .5 0 .4 0 .6 S 0 .6 0 .5 ps-ns 0 .4 0 .3 0 .3 0 .2 0 .2 0 .1 0 .0 MMP-12 0 .1 MMP-3 0 .0 90 100 110120 130 140150 160 170 180190 200210 220 230 240250 260 110 120 130 140 150 160 170 180 190 200 210 220 230 240 250 260 R e sid u e N u m b e r R e s id u e N u m b e r High Rigidity Limited Conformational Flexibility MMP-12 is more rigid than MMP-3. L2/3 L3/4 F157 β3 β5 D158 β1 β4 β2 S225 L229 Apparent Trade-off of Activity and Rigidity in MMP12 for Stability and Flexibility in MMP3 Clemson CI Symposium - Feb 13, 2013 Liang, Bhaskaran et al Biophysical Jl. (2010) Family of NMR structures of STT3P C-ter Domain -the Catalytic Subunit of Oligo Sachcharyl Transferase RCSB PDB code “2lgz” “WWDYG”motif Ost1p binding site CW 110 deg Membrane embedding site / Catalytic center “DK”motif Ave BB rmsd to mean: 0.230 nm Ave heavy atom rmsd to mean : 0.307 nm It is so far the biggest monomeric helical integral membrane protein structure by NMR Clemson CI Symposium - Feb 13, 2013 Huang, Bhaskaran, et al Jl.Biol.Chem. (2012) Structural Gallery - Proteins COBROTOXIN (1COD) TMH0916 (1NR3) CARDIOTOXIN III (2CRS) CARDIOTOXIN II (1CRE) GIP / L-GLUT COMPLEX (2L4T) MMP-12 (2POJ) STT3P (2LGZ) GIP (2L4S) Clemson CI Symposium - Feb 13, 2013 PERTURBATION OF MMP12 BY Triple Helical Peptide AND THE MAPPING OF THEM ON THE STRUCTURE OF MMP12. 0 .0 5 K d = 30 ± 6 M H112 Binding Isotherm G221 0 .0 4 L147 T205 K148 Y240 L224 0 .0 3 H (p p m ) Y113 T210 V243 0 .0 2 0 .0 1 0 .0 0 0 .0 0 0 0 0 .0 0 0 1 0 .0 0 0 2 0 .0 0 0 3 [ a 1 (V )T H P ] (M ) 1 15 Chem ical S hift P ertu rb atio n of H - N N M R Correlatio n S pectru m for Finding Bindin g Site 15 N 1 H Clemson CI Symposium - Feb 13, 2013 Bhaskaran et al. J Biol Chem.(2008) Protein:Ligand Interface Mapping by NMR The NMR paramagnetic surface protection assay correctly predicts the sites of interaction as confirmed by the loss of function mutation studies Clemson CI Symposium - Feb 13, 2013 Palmier, Bhaskaran et al Jl Biol Chem. (2010) EVOLUTIONARY TRACE ANALYSIS Evolutionary Trace Method uses a sequence similarity tree of a family of homologous proteins to highlight residues that are statistically under evolutionary pressure and therefore possess certain functional or structural importance for the family Clemson CI Symposium - Feb 13, 2013 BINDSIght Method - maps of binding sites and distinctive sequence to suggest residues tuning specificity Clemson CI Symposium - Feb 13, 2013 Palmier, Bhaskaran et al. J. Biol. Chem. (2010) CI: Scope and Problems for Interaction Pulse program conversion - Varian and Bruker Consolidation of SW packages on NMR structure determination, dynamics and ligand interactions Develop multiuser remote instrumentation to convert into a state level facility A Virtual Web Based File system for managing NMR Data Rapid structure determination package for the structural genomics proteins Clemson CI Symposium - Feb 13, 2013 BioNMR – External Collaborations Dr. Steven VanDoren, University of Missouri, Columbia, MO : Structural Interactions of Tumor Viral Proteins, E7 and E2F Dr. K.Ramaswamy, UIC, Rockford, IL : Structural Studies of Filarial Proteins Bm-HSP 12.6 Dr. M.Gnanasekar, UIC, Rockford, IL : Structural Studies of Filarial Proteins Bm-TCTP Dr. Peter Myler, Seattle Biomed, Seattle, WA : Structural Genomics of Proteins from Infectious Diseases Dr. Krishna Sharma, University of Missouri, Columbia, MO : Structures of a-Crystallin & Chaperonins of Eye Disease, Cataract Dr. Raghu Kannan, University of Missouri, Columbia, MO : NMR of Gold Nanoparticle Conjugated Bombesin Dr. T. K. S. Kumar, University of Arkansas, Fayetteville, AR : Structural Studies of Fibroblast Growth factor, FGF2 Dr. K. Gunasekaran, University of Madras, Chennai, India : Structural Studies of Filarial Proteins Bm-API Dr. D. Velmurugan, University of Madras, Chennai, India : Structural Genomics of t. thermophilus Proteins Dr. P. Karthe, University of Madras, Chennai, India : NMR structure of Serine glutamate repeat A, a surface adhesin Clemson CI Symposium - Feb 13, 2013 GEAR- CI: SC-Collaborations Dr. John Dawson, University of South Carolina : Identification of Dehaloperoxidase-Substrate Binding Sites Dr. Caryn Outten, University of South Carolina : Structural Studies of iron sensing proteins, Fra2 and Grx Dr. Homayoun Valafar, University of South Carolina : NMR Residual Dipolar Coupling analyses Dr. Nick Grossoheme, Winthrop University: Dr. Heather Evans Anderson, Winthrop University: NMR structure of ForkHead Transcription Factor Protein, FOXO1 Dr. Sondra Berger, University of South Carolina : Dr. Angela Peters, Claflin University : Thymidalate Synthase Mutant (R163K)-Ligand Interactions Dr. Michael Sehorn, Clemson University : Dr. Erin Eaton, Francis Marion University : Dr. Karen Buchmuller, Furman University : Dr. Marcello Forconi, College of Charleston : Dr. Esmaeil Jabbari, University of South Carolina : (CRP) NMR Structural Characterization of vasculogenic peptides Dr. Scott Argraves, University of South Carolina : (CRP) NMR Studies of Fibulins, the extracellular matrix proteins Clemson CI Symposium - Feb 13, 2013 ACKNOWLEDGEMENT Mr. Raghav Nagampalli (Fulbright Scholar) Dr. Angela Peters (Claflin) Dr. Verlie Tisdale (Claflin) School of Natural Sciences and Mathematics, Claflin SC INBRE NASA EPSCoR Space Grant. TRUSTWORTHINESS RESPECT RESPONSIBILITY FAIRNESS CITIZENSHIP CARING Clemson CI Symposium - Feb 13, 2013 THANK YOU ALL Clemson CI Symposium - Feb 13, 2013