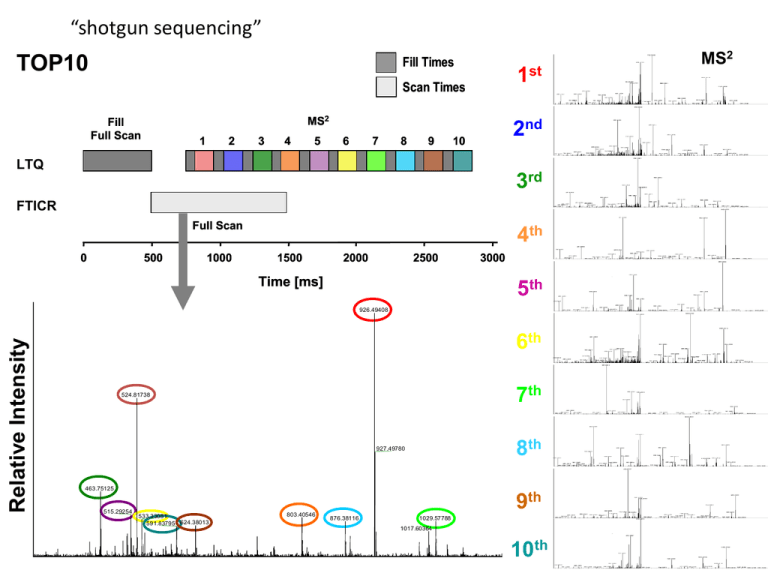
“shotgun sequencing”
TOP10
Fill Times
1st
Scan Times
MS2
Fill
Full Scan
1
2
3
4
5
6
7
8
9
2nd
10
LTQ
3rd
FTICR
Full Scan
0
500
1000
4th
1500
2000
2500
Time [ms]
3000
5th
Relative Intensity
926.49408
6th
7th
524.81738
8th
927.49780
463.75125
515.29254
533.33081
591.83795 624.38013
803.40546
9th
876.38116
1029.57788
1017.60364
10th
MS2
spectral matching
MS/MS Spectrum
0
250
500
750
1000
1250
1500
“shotgun sequencing”
time
“shotgun sequencing”
time
ms1
ms1
ms1
time
ms2
ms2
ms2
distributed spectral matching
6000 spectra x 10s/spectrum = 16 CPU hours
LTQ Orbitrap base peak chromatogram
search time
100
Server
single CPU
Relative Abundance
80
16 hours
60
Server
40
parallel CPUs
20
0
20
30
40
Retention time (min)
50
20 nodes
37 min LC-MS/MS run-time
6186 MS/MS spectra
2308 peptide IDs
(false-positive rate 1%)
287 protein IDs
0.8 hours
sequest
XCorr: goodness of fit
between theoretical
b and y ions from
peptides in the database
dCn: fractional XCorr
difference between the
highest XCorr and next
highest XCorr
yates j.r. 3rd et al. j am soc mass spectrom 5:976-89 (1994)
sequest
time
ms1
ms1
ms1
5000 - 25000
ms2 spectra
time
ms2
2
all ms2 ms
in LC
run
ms2
all ms2 in LC run
all raw
(all ms2 = 1 file)
501.000 (precursor
1001.500
(precursorm/z)
m/z)
+2
+3
1 dta
2
sequest
(charge state)
ms2 array
1 ms2 = 1 file
(all ms2 = ~10000 files)
sequest
all ms2 in LC run
>IPI00000001.2
MSQVQVQVQNPSAALSGSQILNKNQSLLSQPLMSIPSTTSSLPSENAGRPIQNSALPSASITSTSAAAESITPTVELNAL
CMKLGKKPMYKPVDPYSRMQSTYNYNMRGGAYPPRYFYPFPVPPLLYQVELSVGGQQFNGKGKTRQAAKHDAAAKALRIL
QNEPLPERLEVNGRESEEENLNKSEISQVFEIALKRNLPVNFEVARESGPPHMKNFVTKVSVGEFVGEGEGKSKKISKKN
AAIAVLEELKKLPPLPAVERVKPRIKKKTKPIVKPQTSPEYGQGINPISRLAQIQQAKKEKEPEYTLLTERGLPRRREFV
MQVKVGNHTAEGTGTNKKVAKRNAAENMLEILGFKVPQAQPTKPALKSEEKTPIKKPGDGRKVTFFEPGSGDENGTSNKE
DEFRMPYLSHQQLPAGILPMVPEVAQAVGVSQGHHTKDFTRAAPNPAKATVTAMIARELLYGGTSPTAETILKNNISSGH
VPHGPLTRPSEQLDYLSRVQGFQVEYKDFPKNNKNEFVSLINCSSQPPLISHGIGKDVESCHDMAALNILKLLSELDQQS
TEMPRTGNGPMSVCGRC
digest to next peptide
1 dta,
2 dta,
3 dta,
10000
dta
MSQVQVQVQNPSAALSGSQILNK
calculate peptide mass
2426.258812
compare with precursor
peptide mass:
1000.000
3000.000 +/- 1Da
not a candidate
if cand., calc. theoretical spectrum
human
ipi database
correlate, score &
61236
proteins
return
10000
32 xx3,250,000
3,250,000
x3,250,000
3,250,000
times
times
times
times
theoretical “candidate” spectrum
experimental peptide spectrum
correlation
spectrum
-2000
-1500
-1000
-500
0
500
1000
yates j.r.
3rd
1500
2000
et al. j am soc mass spectrom 5:976-89 (1994)
correlation
spectrum
-2000
-1500
-1000
-500
0
500
1000
yates j.r.
3rd
1500
2000
et al. j am soc mass spectrom 5:976-89 (1994)
correlation
spectrum
-2000
-1500
-1000
-500
0
500
1000
yates j.r.
3rd
1500
2000
et al. j am soc mass spectrom 5:976-89 (1994)
similarity scoring
Xcorr score
correlation
spectrum
-2000
-1500
-1000
-500
0
500
1000
yates j.r.
3rd
1500
2000
et al. j am soc mass spectrom 5:976-89 (1994)
similarity scoring – cross-correlation vs dot product
Xcorr score
-1500
-1000
-500
0
500
1000
1500
2000
Dot product
-2000
Dot product
Xcorr (cross-correlation)
non-indexed searching
>ipi00000001.2
1st
MSQVQVQVQNPSAALSGSQILNKNQSLLSQ
PLMSIPSTTSSLPSENAGRPIQNSALPSASITST
SAAAESITPTVELNAL….
1200 +/- 1Da
>ipi00853644.1
61236th
human ipi database
61236 proteins
….AKPNINLITGHLEEPMPNPIDEMTEEQKEY
EAMKLVNMLDKLSREELLKPMGLKPDGTIT
indexed searching
>ipi00001234.11
75 Da
G
>ipi00344567.1
WEFGGHTVLR
1200 +/- 1Da
>ipi00853644.1
20245 Da
human ipi database
61236 proteins
indexed
AKPNINLITGHLEEPMPNPIDEMTEEQEYEA
MLVNMLDLSEELLKPMGLKPDGTITAKPNINL
ITGHLEEPMPNPIDEMTEEQEYEAMLVNML
DLSEELLKPMGLKPDGTIT
scoring & analysis
Score/Metric 1 Score/Metric 2 Score/Metric 3
Peptide A
7.65
0.99
97
Peptide B
6.99
0.87
97
Peptide C
6.21
0.65
97
Peptide D
5.57
0.71
96
Peptide E
3.31
0.44
50
Peptide F
1.85
0.41
41
sensitivity =
precision =
frequency
TP
TN
FN
FP
cutoff/threshold
score/criterion
specificity =
TP
TP + FN
TP
TP + FP
TN
TN + FP
TP + TN
accuracy =
TP + TN + FN + FP
The Results: Distinguishing Right from Wrong
In large proteomics data sets (for which manual data inspection is impossible),
how can we distinguish between correct and incorrect peptide assignments?
Use “decoy” sequences to distract non-peptidic, nonuniquely matchable, or otherwise unmatchable spectra
into a search space that is known a priori to be incorrect
Use the frequency of “decoy” sequences among total
sequences to estimate the overall frequency of wrong answers
(False Positive Rate)
Adjust filtering criteria to achieve a ~ 1% False Positive Rate
Decoy Sequences? A “Reversed” Database!
We generate decoy sequences by reversing each protein sequence in a given
database, such that the resultant in silico digest contains nonsense peptides,
then append the reversed database to the end of the forward database
SEARCHING
Decoy references are
labeled with #
Database searching with SEQUEST occurs from top to bottom – when decoy
references are found, there is an equal probability it could have also mapped to a
non-decoy sequence. So our FPR is (# of decoys) x 2 / total matches.
Target/Decoy Database Searching
Forward database
1. MAGFA→ → →SHTRP
Reversed database
1. PRTHS→ → →AFGAM
Composite Database
Final list
Sequest
Right
F
Wrong (random)
F
R
Unknown
FP
100% 50%50%
Filter
(scoring, mass accuracy, etc)
Generate final list
Estimate FP rate from 2 x Rev (i.e., 4%)
Known
FP
sequest scores: finding true positives
Forward + Reverse
0 .7
0 .7
0 .6
0 .6
0 .5
0 .5
DCn
0 .8
0 .4
0 .4
0 .3
0 .3
0 .2
0 .2
0 .1
0 .1
0
0
0
1
2
3
4
5
6
7
8
0
1
2
3
XCorr
4
5
6
7
XCorr
50
FP
PSM number
DCn
Forward Sequences
0 .8
TP
40
30
20
10
0
0
1
2
3
4
5
6
7
8
XCorr
8
High Mass Accuracy
Mass “Accuracy” in Proteomics:
Precision of mass errors between observed and actual m/z
LTQ FT (SIM)
LTQ Orbitrap &
LTQ FT
AGC target 50,000
to avoid space-charge effects
800
300
600
200
Pept. IDs
Pept IDs
250
150
100
200
50
0
-20
400
-15
-10
-5
0
5
10
15
20
0
-20
-15
-10
-5
0
5
10
Mass accuracy (ppm)
Mass accuracy (ppm)
-0.2 ± 1.0 ppm
0.1 ± 0.4 ppm
15
20
Performance is related to the width of the distribution, not the average error
Haas et al. (2006) Mol. Cell. Proteomics 5, 1326
Olsen et al. (2004) Mol. Cell. Proteomics 3, 608
MMA: True Positives and False Positives
True Positives
False Positives
0
MMA
False positives are distributed evenly across MMA space
50
PSM number
FP
TP
40
30
20
10
0
0
1
2
3
4
5
6
7
8
MS/MS vs MMA: Precision vs Sensitivity
50
PSM number
FP
TP
40
30
20
10
0
0
1
2
3
4
5
6
7
0
8
MMA
MS/MS criteria are strong precision filters – require TP / FP separation for sensitivity
50
40
30
20
10
0
MMA
0
0
1
2
3
4
5
6
7
MMA criteria are weak precision filters – assists MS/MS criteria in improving sensitivity
8
Distracting Wrong from Right: MMA
True Positives
False Positives
0
MMA
Search Space
True Positives
False Positives
Filtered
Filtered
0
Extended Search Space
MMA
Mass Accuracy: Another dimension of selectivity
Forward Sequences
0 .8
0 .7
0 .7
0 .6
0 .6
0 .5
0 .5
DCn
DCn
Tryptic
Search
+/- 2Da
0 .4
0 .3
0 .2
0 .2
0 .1
0 .1
0
0
1
2
3
4
5
6
7
8
0
XCorr
0 .8
1
2
3
0 .7
0 .7
0 .6
0 .6
0 .5
0 .5
0 .4
5
6
7
8
5
6
7
8
0 .4
0 .3
0 .3
0 .2
0 .2
0 .1
0 .1
0
4
XCorr
0 .8
DCn
DCn
0 .4
0 .3
0
Tryptic
Search
+/- 2Da
5ppm
filter
Forward + Reverse
0 .8
0
0
1
2
3
4
XCorr
5
6
7
8
0
1
2
3
4
XCorr
Distracting Wrong from Right: Trypticity
Tryptic Search
True Positives
False Positives
K/R-PeptideK/R-
Partial Enzyme Search
True Positives
Filtered
False Positives
Filtered
A- G- C- S- T- I- L- F- P- M- V- H- D- E- Y- W- Q- N-
K/R-Peptide
PeptideK/R-
A- G- C- S- T- I- L- F- P- M- V- H- D- E- Y- W- Q- N-
What do we have here, hm?
n = 286
dCn
1
0.8
0.6
Unphosphorylated
Phosphorylated
0.4
Reversed Hits
0.2
0
0
2
4
6
8
XCorr
Phosphopeptides: Chemically disadvantaged…
Dataset of phosphorylated and unphosphorylated peptide MS/MS pairs
MSFEILR
P
Singly Phosphorylated (n=207)
Doubly Phosphorylated (n=79)
8
n = 286
XCorr (Phosphorylated)
dCn (Phosphorylated)
1.0
MSFEILR
0.8
0.6
0.4
0.2
0.0
n = 286
6
4
2
0
0.0
0.2
0.4
0.6
0.8
dCn (Unphosphorylated)
1.0
0
2
4
6
XCorr (Unphosphorylated)
8
Phosphopeptides: Less power in XCorr & dCn
XCorr (Ph/UnPh)
2
1.5
Singly
Phosphorylated
1
Doubly
Phosphorylated
0.5
86%
Unphosphorylated
Unphosphorylated
dCn (Ph/UnPh)
0
2
1.5
1
0.5
0
93%
Unphosphorylated
Unphosphorylated
Mass Accuracy: Can it help for phosphorylation?
MS/MS
LTQ
1 2 3 4 5 6 7 8 9 10
0
Yeast Whole-Cell Lysate
1
2
Time (sec)
Red., Alkyl.
SDS-PAGE
Ion Accumulation
for Full MS (1x106)
LTQ
60-80 kDa
3
4
MS/MS
1 2 3 4 5 6 7 8 9 10
Orbitrap
Full MS Scan (R 6x104)
0
Trypsin
IMAC-purification
1
2
Time (sec)
3
4
Mass Accuracy: Rescuing phosphopeptides
SEQUEST partial enzyme search, fully tryptic peptide spectral matches
Orbitrap TOP10
7
LTQ TOP10
n=1311
8
+3: 2.3
7
6
6
+2: 1.3
4
-50
3
0
50
2
XCorr
XCorr
5
5
4
+3: 3.5
+2: 2.7
3
2
1
1
0
0
-750
-500
n=1390
-500
-250
-250
0
250
0
500
750
MMA (ppm)
250
500
750
Mission: Phosphopeptide rescue – accomplished!
1200
1046
0.4% FP
# of phosphopeptides
1000
74%
increase
715
800
600
1.0% FP
1.0% FP
600
400
200
0
LTQ
No MMA
MMA
Orbitrap
search algorithms & phosphorylation
98
sequest
omssa
936
928
Bakalarski et al., Anal. Bioanal. Chem., 2007
phosphorylation site localization
GFDSNQpTWR or GFDpSNQTWR?
Beausoleil et al., Nat. Biotechnol, 2006
phosphorylation site localization
Beausoleil et al., Nat. Biotechnol, 2006
phosphorylation site localization
Taus et al., JPR, 2011
phosphorylation localization rate (FLR)
use non-native phosphoacceptors as “decoys”
Ser + Thr (human proteome): 14.1%
Pro + Glu (human proteome): 14.5%
allow search engine / localization assessment tools
to consider pP and pE as true negative “decoys”
calculate dataset FLR based on frequency of pP + pE “decoys”
Baker et al., MCP, 2011
Chalkey & Clauser, MCP, 2012