PPTX
advertisement

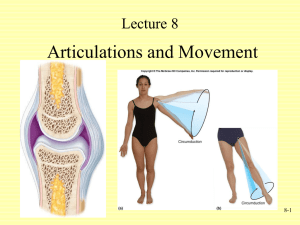
Applications of optical tweezers in protein-protein interaction analysis Ran Yang What are optical tweezers? • Highly focused laser beam holds a dielectric object (e.g. bead) in place using a strong electric field • Use Hooke’s law to estimate the force needed to horizontally displace the bead The Ribosome Modulates Nascent Protein Folding Problem • The transition from ribosome-bound nascent proteins to functional native proteins has only been characterized through computational analysis. • How do proteins attain their native state? • Can we observe their intermediates? Methods • Optical tweezers apply force between the ribosomal subunit and the nascent chain. • T4 lysozyme • Synthesis requires interaction between C and N termini • Added 41aa sequence to C-terminus to allow complete T4 chain to emerge from the ribosome • Apply force to unfold T4 polypeptide then allow refold Results • Protein in solution always refolds correctly, but not the ribosomal-bound T4. • Ribosome-bound protein refolds slower • Increasing the C-extension to 60aa leads to slightly faster refold • Electrostatic interactions between ribosomal surface and charged residues in nascent chain slow down refolding [Fig. D] Results • Folding pathway includes an intermediate that reversible to the unfolded state but irreversible to the native state. [Fig. A] 𝑈 ⇄ 𝐼 → 𝑁 • I is somewhat more stable than U, but N is much more stable than I. • Estimated through force calculations: 3.6pN causes U and I to be equally populated; I is 10nm shorter than U Results • The rate of ribosome-bound I-N transition is much lower than that of the free protein. [Fig. A] • Ribosome-bound U is more compact than free U. • Ribosomal interactions decelerate formation of the native state and stabilizes the intermediate. [Fig. C] Results • If the full polypeptide doesn’t emerge from the ribosome, there is no refolding. • If T4 is fragmented and released from the ribosome, the proteins will fold stably, but they are probably not all functional. [Fig. A, B] • The ribosome may prevent misfolding of incomplete proteins, as a molecular chaperone Conclusions • What is the function of the ribosome with respect to protein transitions from nascent to native state? • Ribosomes slow folding of polypeptide chains that have not been completely synthesized by attracting positively charged residues. • Ribosomes compact polypeptide chains and limits nascent chain interactions. • Ribosomes may complement the activity of other molecular chaperones. ClpX(P) Generates Mechanical Force to Unfold and Translocate Its Protein Substrates Problem • AAA unfoldases degrade damaged polypeptides using ATP hydrolysis to unfold and translocate it to the AAA peptidase chamber. • ClpX is an ATPase that recognizes degradation target via ssrA tag, unfolds target protein, and ports it to the peptidase ClpP, which hydrolyzes polypeptides • By what mechanism does ClpXP unravel the 2’ and 3’ structures of proteins? Methods • ClpXP immobilized on polystyrene beads with X exposed, allowing binding to ssrA • Substrate (GFP) fused to ssrA-tagged titin I27 (red chain) and to dsDNA (blue chain) • Observe ClpX binding to ssrA-tagged substrate when bringing beads close enough together, with ATP • Fixed positions of traps allows observation of ClpX motor force by the movement of the beads Results • Sudden extension followed by retraction of the GFP show unfolding and polypeptide transport respectively. [Fig. B] • Smaller rips are attributed to the polypeptide slipping along the motor. • ClpX pulls in the polypeptide at roughly 8nm/s or about 80aa/s • It seems that GFP unfolds basically all at once (red arrow). The 220aa extension agrees with calculated length of unfolded – folded GFP Results • What if you pull on the beads to create an opposing force? • ClpX stall force is about 20pN, i.e. this is the maximum force ClpX can use to unravel 2’ and 3’ protein structures • Below 13pN, translocation velocity is about constant, suggesting ClpX generates mechanical force and that chemical steps are ratelimiting. [Fig. A] • If you pull even harder, you see the polypeptide translocated in fixed-length steps. [Fig. B] • One rotation of ClpX motor is equivalent to pulling in 1nm of polypeptide Results • There is a short-lived intermediate state when unraveling GFP [Fig. E, red circle] • From the observed lengths of the two “halves” of the rip, we can predict the structure of the intermediate • Residue 130, occurring at the end of a β-sheet is a good candidate [Fig. D, F] Results • Increasing the external force increases the number of pauses during translocation, but not the length of the pauses. • If you slow down the system, it is more likely to pause. • Translocation and pausing could be kinetically competing processes (but why should this be the case?) • Slipping (green circles) after failing to unravel a substrate is most likely caused by temporarily releasing the substrate. • ClpXP complexes are much less prone to slipping, possibly because ClpP digests the polypeptide so that “slipping” would simply cause ClpX to let go of the entire substrate. Conclusions • ClpX can generate enough force to unravel protein substrates. • A motor translocates the polypeptide to ClpP in fixed-length steps (not fixed-aa steps), suggesting that it largely ignores the contours of the substrate itself. • High external forces slow down the ClpX motor, causing more frequent pauses, possibly because ClpX stochastically fails to turnover the next step. • ClpX and ClpXP both form the same intermediate, indicating that unraveling is a function of the substrate, not ClpX. General Conclusions • Optical tweezers allows analysis of forces in protein-protein interactions. • Ribosomal function on nascent polypeptides • Effect of protein motors on polypeptides • Reminder: Must be careful when making assumptions from these data, e.g. what the GFP intermediate looks like based on the length of rips in the folded -> unfolded transition [Fig. 4].