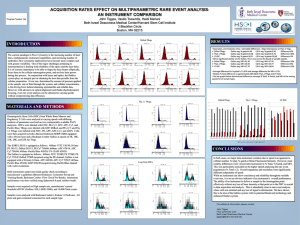
DNA cell cycle analysis is affected both by the type of instrument
advertisement

DNA cell cycle analysis is affected both by the type of instrument used and the binding dye Program No. 161 Ryan Duggan and David Leclerc The University of Chicago Flow Cytometry Facility, Chicago, IL, USA CEN Data Tables (CVs and Ratios) Introduction The choice of a cell cycle dye and instrument are critical elements in a successful cell cycle analysis experiment. Here, we illustrate the predictive potential of using Chicken Erythrocyte Nuclei (CEN) as a tool to choose the most appropriate binding dye and instrument. Also, using a variety of cell cycle dyes and three distinct flow cytometry platforms, a BD LSRII, a Beckman Coulter Gallios and a BD FACSCalibur, we analyzed two parameters, the CV of the G0/G1 population and the G2/G1 ratio, as indicators of the instrument performance. Keeping those factors in mind when planning a cell cycle analysis experiment is key for the acquisition of quality data. Material and Method CENs and the Sp2/0 cell line were stained with a variety of DNA dyes and analyzed on two digital flow cytometers (a BD LSRII and a Coulter Gallios) and an analog instrument (BD FACSCalibur). Data was acquired at a rate of 100-300 events/second. Excitation CEN Singlet Population CV BD BD LSRII BC Gallios FACSCalibur 4.89 4.74 11 PI 7.68 6.13 N/A DAPI 2.16 4.8 N/A DC-Violet 8.32 6.26 8.71 DC-Orange 8.93 8.6 13.9 DC-Ruby 14.2 13.9 25 DRAQ5 DNA Content CEN Singlet/Doublet Ratios (2.0 = theoretical) BC BD LSRII Gallio BD FACSCalibur s 1.98 1.99 2.01 PI 2.01 1.99 N/A DAPI 2 1.99 N/A DC-Violet 2.04 2 2.02 DC-Orange 1.91 2.01 1.95 DC-Ruby 2.02 1.99 1.86 DRAQ5 Sp2/0 Data Tables (CVs and Ratios) DNA Cell Cycle G1 CV BD BD LSRII BC Gallios FACSCalibur 4.6 4.17 3.18 PI 13 15 N/A DAPI 5.32 9 N/A DC-Violet 12 12 10.2 DC-Orange 14 16 15 DC-Ruby 15 16 16 DRAQ5 DNA Cell Cycle G2/G1 Calculated Ratios BD BD LSRII BC Gallios FACSCalibur 1.91 1.96 1.92 PI 1.76 1.8 N/A DAPI 1.79 1.92 N/A DC-Violet 1.62 1.78 1.49 DC-Orange 1.7 1.76 1.6 DC-Ruby 1.58 1.7 1.56 DRAQ5 Sp2/0 example Pics Collection filter BD LSRII BC Gallios BD FACSCalibur BD LSRII BC Gallios BD FACSCalibur DAPI 405 nm 405nm N/A 450/50 450/50 N/A DyeCycle Violet 405 nm 405nm N/A 450/50 450/50 N/A Dye Cycle Green 488nm 488nm 488nm 530/30 530/30 530/30 Dye Cycle Orange 488nm 488nm 488nm 585/42 585/42 585/42 Propidium iodide 488nm 488nm 488nm 585/42 585/42 585/42 DyeCycle Ruby 633nm 633nm 633nm 660/20 660/20 660/20 DRAQ5 633nm 633nm 633nm 660/20 660/20 660/20 • CENs were stained with a final concentration of 5μM of the DyeCycle dyes and 50micrograms per milliliter of PI and DAPI for 30 minutes at room temperature. • Fresh Sp2/0 cells were stained with the DyeCycle dyes and DRAQ5 with a final concentration of 5 μM at 37°C for 30 minutes. • Sp2/0 cells were fixed with 70% iced cold ETOH and stained with a final concentration of 5μM of the DyeCycle dyes and 50micrograms per milliliter of PI and DAPI for 30 minutes at room temperature. CEN example Pics Conclusions • CENs have shown themselves to be useful tools in predicting the quality of the cell cycle analysis. In our hands, the DyeCycle violet and orange provided the best results. This information could potentially save a lot of time and effort when planning a DNA analysis experiment. • As indicators of respectively resolution power and linearity of the detector, the CV of the G0/G1 population and the ratio of the G2/G1 signal can provide valuable insight of the instrument performance. • As for instrumentation, there is certainly some linearity issues associated with our FACSCalibur, however both the newer digital systems, the Gallios and the LSRII showed linear results.