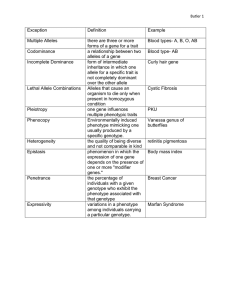
GENETICS: EXERCISES CHAPTER 1 FROM GENE TO PROTEIN 1. Example Gene (DNA): 5’ TCGCC ATG GAA GCA TCA CCC TAG 3’ (positive strand or forward strand) 3’ AGCGG TAC CTT CGT AGT GGG ATC 5’ (negative strand or reverse strand) Alone, the negative strand should be written: 5' CTA GGG TGA TGC TTC CAT GGCGA 3’ messenger RNA: 5’ UCGCC AUG GAA GCA UCA CCC UAG 3’ Protein: Met–Glu–Ala–Ser–Pro–(stop) (= MGASP) In the exercises on gene, if there is no any mention, please consider the provided sequence as a positive strand, direction 5’- 3’. Example on Chargaff’s rule in a double strand DNA: 5’ T T A T A T G C C C A T G G C C G C A G C C G G 3’ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ │ 3’ A A T A T A C G G G T A C C G G C G T C G G C C 5’ 2. Problem 1. This is the sequence of the first exon of a gene (negative strand): 5’ –TCCGCACAGGGTTCCCCAATGCATTTTCCT -3’ Write the complementary DNA sequence (positive strand). Determine the mRNA and peptide sequences encoded by this gene exon. 1 Solution: Positive strand 5’- AGGAAAATGCATTGGGGAACCCTGTGCGGA -3’ - mRNA 5’- AGGAAAAUGCAUUGGGGAACCCUGUGCGGA -3’ - peptide sequence Met-His-Trp-Gly-Thr-Leu-Cys-Gly 2. The following sequence is located at the beginning of a gene. The underlined part corresponds to an intron, which flanks two exons. ATTAGCCATGCTCTCCGTCCCAACTGTAAGTATGCGCGAGATCGTTACCAGGATGATTGT… Determine the sequence of the corresponding mRNA and peptide. Solution - mRNA 5’- AUUAGCCAUGCUCUCCGUCCCAACUGAUGAUUGU -3’ - peptide Met-Leu-Ser-Val-Pro-Thr-Asp-Asp-Cys 3. The following sequence (positive strand) is located in the middle of a bacterial gene: CTCTCCGTCCCAACTGATGATTGT What are the possible peptides corresponding to this sequence? Solution 1. Leu-Ser-Val-Pro-Thr-Asp-Asp-Cys 2. fMet-Ile 4. Write the 3 possible correct codes for the following polypeptides (Multiple answers several codons code for different amino acids), a. Leu – Val – Cys – Lys - Stop 5’- CUUGUAUGCAAAUGA -3’ 5’- CUCGUAUGCAAAUGA -3’ 5’- CUCGUAUGUAAAUGA -3’ b. His – Met – Ser – Thr – Stop 5’- CAUAUGAGUACUUAA -3’ 5’- CAUAUGAGCACUUAA -3’ 5’- CAUAUGAGUACAUAA -3’ 2 5. Here is the sequence of a gene: PROMOTER: TTCCCTAGATAGAGATACTTTGCGCGCACACACATGCAAACGCGCGCAAAAAGG AAAGCCCACCTATAAACTCCAGCCGCAAAGAGAAAACCGGAGCAGCCGCAGCTC EXON 1: ACCTGGCCGCGGGGCGGCGCGCTCGATCTACGCGTCCGGGGCCCCGCGGGGCCGG GCCCGGAGTCGCCATG AAT CGC TGC TGG GCG CTC TTC CTG TCT CTC TGC TGC TAC CTG CGT CTG GTC AGC GCC GAG INTRON 1: GTGAGTTGCCACGGCGGCATGCAGTTGGTTCGCCCCTTTTGGTGTCTGCCCGGCAG EXON 2: GGG ACC CCA TTC CCG AGG AGC TTT ATG AGA TGC TGA GTG ACC ACT CGA TCC GCT CCT TTG ATG ATC TCC AAC GCC TGC TGC ACG GAG ACC CCG GA INTRON 2: GTAAATGGAATCCTCGCCCCGCGCTCCGGCCCTCCGAGGAGGTGGGG CCGCCTGGTGTCTGACTGTGACTTCTCCTGCAG EXON3: A GGA AGA TGG GGC CGA GTT GGA CCT GAA CAT GAC CCG CTC CCA CTC TGG AGG CGA GCT GGA GAG CTT GGC TCG TGG AAG AAGGAGCCTGG a) Where does transcription start? At the first nucleotide of exon 1 b) What is the sequence of the messenger RNA precursor and of the mature messenger RNA? Precursor : exon1+intron1+exon2+intron2+exon3 Mature: exon1+exon2+exon3 (Replacing T by U) c) Where does translation start and what is the peptide sequence (first ten residues)? Asn-Arg-Cys-Trp-Ala-Leu-Phe-Leu-Ser-Leu d) Where does translation stop? TGA in exon 2 6. A somatic cell that has 2n=24 chromosomes undergo mitosis 5 times continously. Please calculate: - The number of cells after the whole process? 25 - The number of chromosomes that the environment did provide to the whole process? 24*(25-1) 7. Complete the following table: Nucleotide Components and Function Nucleic Acid Type DNA deoxyribose Name of the sugar present in nucleotides mRNA ribose 3 tRNA ribose Name of bases present in nucleotides Function Where can you find each of these in a eukaryotic cell? Guanine Cytosine Thymine Adenine Store genetic material of organism Nucleus Mitochondria Chloroplast Guanine Cytosine Uracil Adenine Directs aa sequence of protein Guanine Cytosine Uracil Adenine Transport aa to site of protein synthesis Nucleus Mitochondria Chloroplast Cytosol (free ribosome) ER rough Nucleus Mitochondria Chloroplast Cytosol(free ribosome) ER rough Annex Figure 1.The genetic code. A Alanine Ala I Isoleucine Ile C Cysteine Cys K Lysine Lys D Aspartate Asp L Leucine Leu M Methionine Met E (aspartic acid) Glutamate N Asparagine Asn P Proline Pro F (glutamic acid) Phenylalanine Phe Q Glutamine Gln G Glycine Gly R Arginine Arg H Histidine His S Serine Ser Glu 4 T Threonine Thr V Valine Val W Tryptophan Trp Y Tyrosine Tyr Figure 2.Amino acid names and abbreviations. 5 CHAPTER 2 GENE VARIANTS &POLYMORPHISMS Problems 1. This is the sequence of the first exon of a mouse gene. CGGGCACC ATG AGC GAC GTG GCT ATT GTG… a) What would be the consequence of deleting the boxed nucleotides? Please write down the protein sequences before and after the mutation. Solution: Deleting the boxed nucleotides results in a frame-shift that changes the reading of subsequent codons, so it will alters the entire amino acid sequence. Before: Met-Ser-Asp-Val-Ala-Ile-Val After: Met-Ser-Arg-Gly-Tyr-Cys b) What would be the expected impact of this deletion on the protein function? The protein might loss its function because its function associates with its high-ordered structure which depends on the sequence of amino acid. This often disrupts the biochemistry process in the cell. 2. The following sequence is located at the beginning of a gene. The underlined part corresponds to an intron, which flanks two exons. ATTAGCCATGCTCTCCGTCCCAACTGTAAGTATGCGCGAGATCGTTACCAGGATGATTGT… What would be the expected impact of the following mutations: a) ATTAGCCATGCTGTCCGTCCCAACTGTAAGTATGCGCGAGATCGTTACCAGGATGAT… C to G There is no change in the aa sequence because both codons CUG and CUC coded for Leu b) ATTAGCCATGCTCTCCGTCCCAACTGTAAGTATGAACGAGATCGTTACCAGGATGAT… CG to AA There is no change in aa sequence because the mature mRNA doesn’t consist of intron. c) ATTGGCCATGCTCTCCGTCCCAACTGTAAGTATGCGCGAGATCGTTACCAGGATGAT… A to G 6 There is no change in aa sequence because the mRNA is translated from the first ATG codon which stands after the mutation. 3. A research team sequenced a human gene and the corresponding mRNA. Here is the sequence of the genomic DNA. The parts that are identical between the genomic DNA and the mRNA are written using uppercase letters. agcgaaatttaatgagcgtgtaacaggggactgaaaatcctgatttctcaAGCTATCAAA del 1 GGTTTATAAAGCCAATATCTGGGAAAGAGAAAACCGTGAGACTTCCAGATCTTCTCTGGT GAAGTGTGTTTCCTGCAACGATCACGAACATAAACATCAAAGGATCGCCATGGAAAGgta del 2 agtgtgacaactcactgcgttggtggctcgcgttcttatgagctaagGGTCCCTCCTGCT GCTGCTGGTGTCAAACCTGCTCCTGTGCCAGAGCGTGACCCCCTTGCCCATCTGTCCCGG Del 3 CGGGGCTGCCCGATGCCAGGTGACCCTTCGAGACCTGTTTGACCGCGCCGTCGTCCTGTC CCACTACATCCATAACCTCTCCTCAGAAATGTTCAGCGAATTCgtaagtaccatgcttct ggcttcctattgaatttgtctcatcatttccagGATAAACGGTATACCCATGGCCGGGGG del 4 TTCATTACCAAGGCCATCAACAGCTGCCACACTTCTTCCCTTGCCACCCCCGAAGACAAGGA GCAAGCCCAACAGATGAATgtgagtccttcatccaggctttgca a) Which parts of that sequpromoterence correspond to exons, introns and promoter? b) What is the sequence of the peptide encoded by this gene (first ten residues)? c) What will be the impact of each of the deletions indicated by boxed nucleotides on the protein sequence and length? Solution: a) Promoter: first part with lowercase letter Exons: all parts with the uppercase letter Introns: all parts with the lowercase letter except the first one b) Met-Glu-Arg-Val-Pro-Pro-Ala-Ala-Ala-Gly c) - del 1: May disrupt the normal processes of gene activation by disturbing the ordered recruitment of transcription factors at the promoter. - del 2 : The mutation happens before the start codon, so it does not affect the coded message. As a result, the related products do not change. - del 3: Deleting the boxed nucleotides results in a frame-shift that changes the reading of subsequent codons, so it will alters the entire amino acid sequence. The polypeptide length (shorter) and chemical composition will be changed, resulting in a non-functional protein that often disrupts the biochemical processes of a cell. 8 - del 4 : Mutation in introns: It does not cause change in the coded message, but may result in a defect in RNA splicing and so affect the corresponding protein. Intronic mutation however can introduce novel splice sites, activate novel promoters (which may direct sense or antisense transcription causing alterations in mRNA, miRNA or lncRNA expression), or introduce/eliminate enhancer activity, so these could affect gene expression levels (affect regulatory region) 4. a) Gene B has 390 Guanine and the total number of hydrogen bonds is 1670, is substitution mutated in 1 pair of nucleotides to gene b. Gene b has 1 hydrogen bond more than gene B. Calculate each type of nucleotide in gene b. Solution: - In allele B 7 The number of Cytosine = Guanine= 390 The number of Thymine = Adenine =(1670-390*3):2=250 - In allele b There is 1 hydrogen bond more than allele B. Therefore, the mutation in 1 pair of nucleotide is the substitution of 1 pair A-T into 1 pair G-C The number of C=G=390+1=391 The number of T=A=250-1=249 b) Gene X has 3600 hydrogen bonds and the number of nucleotide Adenine is equal to 30% the number nucleotide of the whole gene. Gene X is mutated type deletion 1 pair of nucletode A-T to gene x. A cell with heterozygote genotype Xx undergoes mitosis to 2 daughter cells. Please calculate each type of nucleotide that the environment need to provide to the process. Solution: - In allele X: We have + The number of hydrogen bonds=2*A+3*G=3600 + %A=%T=30% => %G=%C=20% The number of A=T=900 The number of G=X=600 -In allele x: A=T=900-1=899 G=C=600 The number of Nu provided by the environment is 600+900+899+600=2999 5. a) A gene D has 4800 hydrogen bonds and the ratio of nucleotide number Adenine/Guanine = 1/2, , is substitution mutated in 1 pair of nucleotides to gene d with 4801 hydrogen bonds. Calculate each type of nucleotide in gene D and d. Solution: -In allele D A=T=600 G=C=1200 -In allele d A=T=600-1=599 G=C=1200+1=1201 b) Gene A with 3000 hydrogen bonds and the number of nucleotide Adenine is equal to Guanine, is mutated to gene a. When gene a underwent DNA replication, the environment provided 2398 nucleotides. Which kind of mutation that gene A had? Solution: - In allele A: A=T=600 G=C=600 - In allele a: The number of nucleotides provided by environment after 1 time of DNA replication is equal to the number of nucleotide of allele a Therefore, the allele a has loss 2 nucleotide which means the mutation is deletion. 6. This is the result of a paternity test of an alleged father (Jim Doe) and child (John). Please check if Jim is biological father of John, in case yes, point out which allele the child inherited from the father. 8 Solution: Jim is the biological father of John 7. Using the genetic code table (attached), fill in the amino acid sequence starting with the first ATG of the coding strand. It helps to mark off the codons by threes. Y 14 16.3 or 17.3 17 9 10 or 16 12 13 23 10 9 6 15 29 or 30 11 11 9 or 11 13 or 14 18 14 18 DNA (5')G G A T A G C A T G A A A C C C G C A T A A (3') amino acid : Met-Lys-Pro-Ala How would the amino acid sequence above change with the following changes (mutations) in the DNA code (changes are marked in bold-face): a. (5')G G A T A G C A T G A A A C C A G C A T A A (3') change to T amino acid: Met-Lys-Pro-Ala b. (5')G G A T A G C A T G A A A C C C C C A T A A (3') change to A amino acid: Met-Lys-Thr-Ala c. (5')G G A T A G C A T G A A A - C C G C A T A A (3') delete amino acid : Met-Lys- Pro-His d. (5')G G A T A G C A T G T A A C C C G C A T A A (3') change to C amino acid : Met-Gln-Pro-Ala 9 8. A diploid organism with 4000 spermatocytes undergoes meiosis to produce gametes. During meiosis there are 40 cells with 1 pair of unseparated chromosomes in meiosis I, meiosis II took place normally, the rest of the cells meiosis normally. Please indicate: a) What is the percentage of non-mutated gametes? 90% b) What percentage of mutant gametes that have more 1 chromosome than normal? 5% c) What percentage of mutant gametes that have less 1 chromosome than normal? 5% Solution: a) normal gametes : (4000-40) x 4= 15 840 percentage of non-mutated gametes: 15 840/ 16 000 = 99% b) after meiosis II, each mutated cell produces 2 gametes that have more 1 chromosome than normal => 40 x 2 = 80 gametes that have more 1 chromosome than normal => 80/ 16 000 = 0.5% c) after meiosis II, each mutated cell produces 2 gametes that have LESS 1 chromosome than normal => 40 x 2 = 80 gametes that have less 1 chromosome than normal => 80/ 16 000 = 0.5% 9. In Drosophila (2n=8), an individual has two chromosomal structure mutations: a chromosome in the 1st pair carries deletion mutation, and a chromosome in the 3rd pair carries inversion mutation. If the meiosis process takes place normally, please calculate: a. Percentage of the normal gametes? 1/4 b. Percentage of the gametes carry the deletion chromosome? 1/2 c. Percentage of the gametes carry both the deletion chromosome and the inversion chromosome? ¼ Solution: We have 2n = 8 => n=4 => There are four pairs of chromosomes with drosophila. Assume each different chromosome by an alphabet letter So we have 1st pair is A//a 2nd pair B//b 3rd pair D//d 4th pair E//e Assume that A is the deletion one D is the inversion one. a. Percentage of the normal gametes? [1(a) x 2(B/b) x 1(d) x 2(E/e) ]/[2(A/a) x 2(B/b) x 2(D/d) x 2(E/e)] = 1/4 b. Percentage of the gametes carry the deletion chromosome? [1(A) x 2(B/b) x 2(D/d) x 2(E/e) ]/[2(A/a) x 2(B/b) x 2(D/d) x 2(E/e)] = 1/2 c. Percentage of the gametes carry both the deletion chromosome and the inversion chromosome? [1(A) x 2(B/b) x 1(D) x 2(E/e) ]/[2(A/a) x 2(B/b) x 2(D/d) x 2(E/e)] = 1/4 10 CHAPTER 3 MENDEL’S LAWS Problems 1. Here are four pedig rees. The black symbols rep resent patients suffering from a rare disease that is transmitted according to Mendel’s law. a) For each family, determine whether 1, recessive because the normal parents have the trait is dominant or recessive. b) Determine the genotypes of the affected individuals and their parents. affected child Child : aa Each parent: Aa 2, dominant because the affected parents have normal child Affected children in second generation: Aa because one of their parents is normal (aa) and the affected parent have genotype Aa because they have normal child The 3 middle affected children in third generation have genotype either AA or Aa because their parents are all affected and have genotype Aa(since one of their children is normal) The last affected child in the third generation :Aa because he has normal children 3. recessive because the normal parents have affected children parents: Aa affected child: aa 4. same as 3 11 2. Crossing two drosophila flies (both) with normal wings produces 27 individuals with short wingsand 79 with normal wings. a) How is the short wings trait transmitted? b) What are the parents genotypes? c) If flies with short wings are crossed with one of the parent flies, how many normal flies are expected in an offspring of 120 flies? Solution: a. Short wings : normal wings=1:3 And the parents are all normal wings the short wings trait is recessive and transmitted according to Melden’s law b. A: dominant, a: recessive Parents: AaxAa c. Short wings: aa Aaxaa => 1Aa:1aa Normal flies in 120 offsprings: ½ x 120=60 3. Albinism in plants is the inability to make chlorophyll. It is a recessive trait. In an experiment, heterozygous tobacco plants undergo auto-fertilization and produce 600 seeds, of which 80% germinate. a) How many albino plants will be obtained? b) How many plants will show the parental genotype? Solution: a. Assume that dominant allele is A; recessive allele is a. Heterozygous parents Aa x Aa => ¼(AA) : 2/4(Aa) : ¼(aa) => ¾ normal : ¼ albinism. Albino seeds: 600 x ¼ = 150 (seeds) Albino plants: 150 x 80% = 120 ( plants) b. Parental genotype seeds: 600 x ½ = 300 (seeds) Parental genotype plants: 300 x 80% = 240 (plants) 4. The ability to taste phenylthiourea (or phenylthiocarbamide) is a genetic trait that is transmitted according to Mendel’s laws. Tasters are able to recognize a solution containing only 0.005% of this product, by contrast to non-tasters, who are not able to detect this molecule even at much higher concentrations. Two parents who are both able to taste phenylthiourea have four children, of whom two are unable to taste the molecule. a) How is this trait transmitted? b) What are the parents’ genotypes? c) If this couple has a fifth child, what is the probability that the child is able to taste phenylthiourea? 12 Solution: Two parents who are both able to taste phenylthiourea have four children, of whom two are unable to taste the molecule. Able to taste x Able to taste => Unable to taste => Able to taste is Dominant; Unable to taste is recessive; Assume that dominant allele is A; recessive allele is => Parents : Aa x Aa. a) Able to taste is Dominant; Unable to taste is recessive b) Aa x Aa. c) Aa x Aa => ¼(AA) : 2/4(Aa) : ¼(aa) => ¾ able to taste : ¼ unable to taste. => 3/4 is the probability that the child is able to taste phenylthiourea 5. Normal human skin pigmentation is dominant over the albino trait. If an albino man expects a child with a woman who has normal skin pigmentation and whose father was albino, what are the possible ge notypes and phenotypes of the child? Solution: Assume that dominant allele is A; recessive allele is a. The albino man : aa The normal woman has normal skin pigmentation whose father was albino is received one ‘a’ allele from her father => Her genotype: Aa => Aa x aa => ½(Aa) : ½(aa) => ½ normal : ½ albino. 6. In this pedigree, patient represented by a black symbol are born deaf. a) What can be concluded in term of transmission of this trait? b) What are the possible genotypes of individual III2? Solution: a) I3 normal x I4 normal => II3 deaf => Normal is dominant ; Deaf is recessive. Assume that dominant allele is A; recessive allele is a. b) II3: aa. III2 is normal but received one ‘a’ allele from the II3 father => Aa. 7. The dominant allele A in Ayrhirecows produces a cut in the ears. In the following pedigree, in which black symbols represent individuals with cut ears, determine thefrequency of this trait in the offspring from the following matings: (a) III1 X III3 (b) III2 X III3 (c) III3 X III4 (d) III1 X III5 (e) III2 X III5 13 Solution All the normal individuals in the pedigree have the genotype : aa. III1 : aa ; III2: Aa ( Because receive one allele ‘a’ from II1); III3: aa; III4: aa; III5: Aa ( Because receive one allele ‘a’ from II6) (a) III1 X III3 : aa x aa (b) III2 X III3 : Aa x aa (c) III3 X III4 : aa x aa (d) III1 X III5 : Aa x aa (e) III2 X III5 : Aa x Aa 8. In Linumusitatissimum (flax, used to make linen textile), the blue color of the flowers (b) and the long fiber length (l) are recessive traits. A farmer mate true breeding plants with short fibers and white flowers with plants with blue flowers and long fibers. a) What are the genotypes of these plants? b) What is the geno type of the F1generation? c) If plants from F1 are auto-fertilized and 800 F2 plants are obtained, 1. calculate the expected number of plants with white flowers and long fibers 2. determine the percentage of the different genotypes among these (white flowers and long fibers). Solution: a) Short fibers and white flowers => Dominant , dominant and this is true breeding plants => BBLL. Blue and long fibers => Recessive , Recessive => bbll. b)P: BBLL x bbll => F1: BbLl c) If plants from F1 are auto-fertilized and 800 F2 plants are obtained, F1:BbLl x BbLl => (3 white : 1 blue) x ( 3 short : 1 long) => (9 white short : 3 white long : 3 blue short : 1 blue long) 1. calculate the expected number of plants with white flowers and long fibers : 3/16 x 800 = 150 ( plants) 2. determine the percentage of the different genotypes among these (white flowers and long fibers): White long genotypes : BBll and Bbll => We need to compare the proportion of BB and Bb => We have Bb x Bb => F2 genotypes proportion : (1BB:2Bb:1bb) => BB:Bb=1/2 => 1BBll : 2Bbll 9. In a bovine population, two co-dominant alleles control the coat color: R (red) and r (white). Heterozygous individuals present an intermediate color named “rouan”. The “strait hair” 14 trait is dominant over the “curly” trait (s). What is the result of mating a red curly bull with a homozygous white cow with strait hair? a) What is the offspring phenotype? b) What is the offspring and parents genotype? c) What is the result of mating a F1individual with a rouan curly bull? Solution: RRss x rrSS => RrSs ( Rouan, strait) a) Rouan, strait hair. b) Offspring: RrSs ; parents : RRss x rrSS. c) F1: RrSs x Rrss ( Nhân độc lập từng kiểu hình của từng tính trạng với kiểu hình của tính trạng còn lại sẽ được kiểu hình của offsprings) =>F2: (1 Red : 2 Rouan : 1 White) x ( 1 Strait : 1 Curly) => ( 1 Red Strait : 1 Red Curly : 2 Rouan Strait : 2 Rouan Curly : 1 White Strait : 1 White Curly) 10. Short hair is a dominant trait in guinea pigs and is controlled by a single gene (allele L), while the allele l corresponds to long hair. The fur color is controlled by a gene which has two co-dominant alleles, such that: the YY genotype = yellow, YW = cream and WW = white. If individuals that have the genotype Ll YW are mated, what will be the possible phenotypes in the F1 generation and the percentages? Solution: Ll YW x Ll YW => (1LL:2Ll:1ll) x (1YY : 2YW : 1WW) => (1 LLYY : 2LLYW : 1LLWW : 2LlYY : 4LlYW : 2LlWW : 1llYY : 2llYW : 1llWW) = > (3 Short Yellow : 6 Short Cream : 3 Short White : 1 Long Yellow : 2 Long Cream : 1 Long White) 11. A man with Huntington disease (autosomal dominant – rare diseases) and the AB blood group married a healthy woman who is O. The genes are independent. What is the probability that their child is healthy and A? Solution: We have Huntington disease is a rare disease, so we can assume that the man carried that disease have heterozygous genotype . Denote : H – Huntington h – normal Ia – Blood type A Ib – Blood type B Io – Blood type O The man genotype : HhIaIb The healthy woman genotype : hhIoIo The child is healthy and A must have hhIaIo genotype => The probability is: ½(hh)x1/2(IaIo)=1/4. 15 12. Two male drosophila flies (1 & 2) are mated with two females (3 & 4). These parents have long wings and a grey body.The results of the mating are: 1 x 3 : 153 grey individualswith long wings 58 grey individuals with wing remnants 1 x 4 : 26 black individuals with long wings 73 grey individuals with long wings 2 x 3 : 112 grey individuals with long wings 2 x 4 : 89 grey individuals with long wings What are the parent genotypes? Solution: All parents have long wings and grey body producing wing remnant and black individuals The dominant alleles: A : Grey B: Long The recessive alleles: a: Black b: remnants - The color of body trait: 1x4: grey : black=3:1=> 1 and 4 : Aa 1x3: 100% grey=> 3 is AA 2x4:100% grey=> 2 is AA - The wings trait: 1x3: long : remnants = 3:1 => 1 and 3 : Bb 1x4: 100% long => 4 is BB 2x3: 100% long=> 2 is BB 1 is AaBb /2 is AABB/ 3 is AABb/ 4 is AaBB 16 CHAPTER 4 PARTICULAR CASES INCLUDING CROSSING OVER Problems 1. A farmer crosses two true breeding types of peonies: one has red flowers with large leaves (R and G dominant alleles, respectively), the other is white with small leaves. a) If these two traits are controlled by two genes, what will be the phenotype of the F1 offspring? b) If F1 individuals undergo auto-fertilization, what will be the percentages of each phenotype in F2 if the two genes are independent? c) Same if the two genes are tightly linked? d) Same if the two genes are separated by a distance of 10 Morgan units (centimorgan)? Solution: a. Consider the flower color trait , true breeding type cross with each other : RR (red flower ) x rr ( white flower ) => Rr ( red flower ) Consider the leaves trait , true breeding type cross with each other : GG (large leaves) x rr (small leaves) => Gg ( large leaves ) => Combine those two traits, we have the F1 phenotype : Red flower, large leaves. b. The genes are independent => F1 genotype : RrGg F1 goes auto-fertilization => F1 x F1 : RrGg x RrGg Gametes F1: ( 1RG : 1Rg : 1rG : 1rg ) x ( 1RG : 1Rg : 1rG : 1 rg ) F2 Genotypes: 1 RRGG: 2RRGg: 1 RRgg : 2RrGG: 4 RrGg: 2Rrgg: 1:rrGG: 2rrGg: 1rrgg F2 Phenotypes: 9 red large : 3 red small : 3 white large : 1 white small c. P true breeding : RG // RG x rg // rg Gametes P : (1RG) x (1rg) F1 genotype : RG // rg F1 goes auto-fertilization => F1 x F1 : RG // rg x RG // rg Gametes F1: (1RG : 1 rg) x (1RG : 1 rg) F2 genotype : 1RG//RG: 2RG//rg: 1rg//rg F2 phenotype : 3 red large : 1 white small d. We have two genes are separated by a distance of 10 Morgan units => The recombination frequency is 10%. F1 genotype : RG // rg F1 goes auto-fertilization => F1 x F1 : RG // rg x RG // rg Gametes F1: (45%RG : 45%rg : 5%Rg: 5%rG) x (45%RG : 45%rg : 5%Rg: 5%rG) F2 phenotype : 70.25% Red large : 4.75% Red small : 4.75% White large : 20.25% White small 17 2. The success of renal transplantation depends on three human histocompatibility genes, HLAA, HLA-B and HLA-C, which must match between the donor and the receiver. A single mismatch may cause the kidney rejection. Each gene has multiple co-dominant alleles. These three genes are located very close to each other on chromosome 6, so that the recombination rate is very low (below 1%). The father has the following genotype: A1, A2, B24, B10, Cw4 and Cw7 and the mother is A1, A1, B11, B7, Cw5 and Cw8. Their first boy is A1, A1, B24, B11, Cw7 and Cw8. What is the probability that the second child is compatible with his/her brother? Solution: The first boy received A1 from both father and mother , received B24 and Cw7 from father, received B11 and Cw5 from mother. => The first child genotype : A1B24Cw7 // A1B11Cw8. => The father genotype : A1B24Cw7 // A2B10Cw4. => The mother genotype : A1B11Cw8 // A1B7Cw5. We need the second child compatible with his/her brother => The second child genotype A1B24Cw7 // A1B11Cw8. The probability is : 50% x 50% = 25% 3. Wild type drosophila are mated with mutants with yellow eyes (s phenotype) and no antenna (bobbed or B phenotype). F1 offspring have wild type eyes and “bobbed” antennas. When F1 individuals are mated, the F2 result is the following: 101 individuals are 32 individuals are 31 individuals are 13 individuals are [wt B] [wt wt] [s B] [s wt] a) Are these genes linked? b) What are the genotypes of the parents and the F1 individuals? Solution: a) eye’s color: wt/s = (101+32)/(31+13) = (3:1) antenna present: B/wt = (101+31)/(32+13) = (3:1) ( 3wt : 1s ) x (3B : 1wt) = 9wtB : 3wt wt : 3sB : 1swt → independent → no linked b) Because:The ratio of wt/s=3:1 => wt is dominant => wt : A , yellow: a The ratio of B/wt=3:1 => Bobbed is dominant => Bobbed: B, wt: b => P: AAbb x aaBB (wild, wild ) ( yellow, bobbed) F1: AaBb ( wild,bobbed) 4. A man inherited cataract from his mother (normal fingers and toes) and polydactyly from his father (normal eyes). The two diseases are caused by dominant alleles of 18 genes that are tightly linked. This man marries a healthy woman. What will be the phenotype of the children? Solution: The two diseases are caused by dominant alleles => A: cataract, a: NO cataract B: polydactyly, b: NO polydactyly Man inherited from mother ( Ab) and from father ( aB) => Man : Ab/aB Man + woman ( healthy) -> Ab/aB + ab -> children : ½ Ab ½ aB => ½ cataract ½ polydactyly 5. Here is a map of the locus for the a, b and c genes: a/A b/B c/C 10 cM 15 cM What will be the percentages of gametes produced by an individual who is A-b-C ? a-B-c Solution: The double recombination frequency is : 0.1 x 0.15 = 0.015 => The double recombination gametes are : A-B-C = a-b-c = 0.015 /2 = 0.0075 The single recombination gametes at A are : a-b-C = A-B-c = (0.1 - 0.015 ) /2 =0.0425 The single recombination gametes at C are : A-b-c = a-B-C = ( 0.15 - 0.015 ) /2 =0.0675 The normal gametes are : A-b-C=a-B-c= 0.5 - 0.0075 - 0.0425 - 0.0675 = 0.3825 6. Two types of drosophila are studied: Wild type flies: mutants: red eye (red) Grey body (grey) Normal wings (norm) chestnut eye (ch) black body (bl) indented wings (ind) Wild type males are mated with mutant females.The F1 population is wild type.Then, a) a F1 female is mated with a mutant male (offspring = F2a) b) a F1 male is mated with a mutant female (offspring = F2b) 19 Here are the phenotypes of the two populations F2a and F2b: 1. F2a Eye Body Wings red grey norm red grey ind red bl norm Red Bl Ind Ch Grey Norm ch grey ind Ch Bl norm ch bl ind 616 65 170 147 144 172 66 620 red grey norm ch bl ind 542 551 2. F2b eye body wings a) How would you explain the F2b results? b) Determine the genetic map of the locus. Solution: P : Wild types x mutants => 100% wild type => Wild type of 3 genes are all dominant and F1 is heterozygous in all genes. Denote : A - red eye >> a - chestnut eye B - Grey body >> b - black body D - Normal wings >> d - indented wings . If the genes are independent, the ratio of phenotype in F2a and F2b should be similar => The genes are not independent => The genes are linked. => F1 genotype : ABD // abd The mutant phenotype ( both female and male ) : abd // abd a. We know that, the male drosophila do not have the gene interchange => Mating B : male ABD // abd x female abd // abd Gamete F1B : (1 ABD : 1 abd ) x (1 abd ) F2B genotypes : 1 ABD//abd : 1 abd//abd F2b phenotypes : 1 red,grey,normal : 1 chestnut,black,indented. => AS THE RESULT IN REALITY. b. We know that, the female drosophila occurs gene interchange . We have to find the double recombination phenotype in the table The double recombination frequency is the product of two recombination frequencies , so it’s phenotype will most likely be the smallest, these are : %Aa,Bb,dd = %aa,bb,Dd=> The double recombination gamete that F1 female create is : ABd and abD => The middle gene is D/d. The distance between A/a and D/d is : (65 + 66 + 144 + 147) / 2000 = 21.1 cM The distance between B/b and D/d is : ( 65 + 66 + 170 + 172 ) / 2000= 23.65 cM 7. In experiments using drosophila confuse, researchers observed that the vermillon trait (v allele) is responsible for dark red eyes. If homozygous a wild type female (light red eyes) is mated with a vermillon male (mating A), the offspring includes 61 wild type females and 57 20 wild type males. If a vermillon female is mated with a wild type male (mating B), the offspring is 88 wild type females and 79 vermillon males. a) How would you interpret these results? b) What are the parents and F1genotypes? c) If F1individuals are mated, what phenotypes and percentages will be observed in F2? Solution: a) We have that in mating A, homozygous wild type female mated with vermillion male => 100% wild type => Wild type is dominant against vermillion. Also in mating B , the recessive trait ( vermillion) in mating B only occurs in males => This trait is a X-linked trait. b) Denote that : wild type : V V>>v mating A P : XV XV x Xv Y Gametes P : ( 1 XV ) x (1Xv :1 Y) F1 genotypes: 1 XV X v :1 XVYmating B P: Xv Xv x XV Y Gametes P : ( 1 Xv ) x (1XV :1 Y) F1 genotypes: 1 XV X v :1 XvY c) If F1 Individuals are mated, what phenotypes and percentages will be observed in F2? mating A’ F1: XVXv x XVY Gametes F1: ( 1XV : 1Xv ) x ( 1XV :1Y ) F2 genotype: 1XVXV : 1 XVXv : 1XVY: 1XvY => F2 phenotype : 2 normal female : 1 normal male : 1 vermillon male mating B’ F1: XVXv x XvY Gametes F1: ( 1XV : 1Xv ) x ( 1Xv :1Y ) F2 genotype: 1XVXv : 1 XvXv : 1XVY: 1XvY => F2 phenotype : 1 normal female : 1 vermillon female : 1 normal male : 1 vermillon male 8. A healthy woman, whose father died from hemophilia (X-linked recessive), married a healthy man. Could their children suffer from hemophilia? Solution: Heathy woman has died father from hemophilia (X-linked recessive) Hemophilia is recessive: a=> normal A =>Her father genotype: XaY A healthy woman: XAXa Her healthy husband: XAY => XAX a x XAY Gametes: (1XA : 1Xa) x (1XA : 1Y) Children phenotypes: 2 Healthy female : 1 healthy male: 1 hemophilia male => their children could suffer from hemophilia with probability P=1/4 21 CHAPTER 5 POPULATION GENETICS Problems 1. In the course of a study dealing with blood group frequencies in a population of 2000 individuals, part of the results were lost. We know that 800 individuals were O and 780 were A. a) What are the allelic frequencies? b) What are the phenotype frequencies? Solution: a) Assume that allele frequency of IA = p Assume that allele frequency of IB = q Assume that allele frequency of IO = r There are 800 blood O individuals => IOIO = 800 / 2000 = 0.4 √10 => r^2 = 0.4 => r = 5 There are 780 blood A individuals => IAIO + IAIA = 780 / 2000 = 0.39 => p^2 + 2.p.r = 0.39 => p = q=1–p–r= b) Blood A : 0.39 Blood O : 0.4 √79−2√10 10 10−√79 10 √79−2√10 10−√79 x 10 10 10−√79 √10 10−√79 Blood B : ( 10 )2+2x 5 x 10 Blood AB : 2x 2. A. If the frequency of albino (recessive trait) in a given population is 0.0009, what will be the frequency of heterozygous individuals? B. Huntington disease (autosomal dominant) has a frequency of 1 case in 25 000. Calculate the frequency of the mutated allele in the population. Solution: A. The allelic frequency of albino = √0.009= 0.03 => The frequency of heterozygous individuals = 2x0.03x(1-0.03)=5.82% 1 B. The % of normal allele=√1 − 25000=99.99% The % of mutated allele=100%-99.99%=0.01% 22 3. A. If, in 5% of men are color blind (recessive X-linked trait) in a given population, what will be the frequency of color blind women? B. If allele frequencies corresponding to the blood group gene was: p(A) = 0.3 ; q(B) = 0.1 ; r(o) = 0.6, what will be the phenotype percentages? Solution: Assume that : XA : normal ; Xa : Color blind XX: p2 (XAXA) + 2pq(XAXa ) + q2 (XaXa ) = 1 XY: p(XAY) + q(XaY) = 1 => Xa = 0.05 ; XA = 1-0.05 = 0.95 => The frequency of color blind women : q^2 = 0.05^2 = 0.25% 4. A. The ability to taste phenylthiocarbamide is a dominant trait in humans controlled by allele G of a gene. In a given population, 2800 individuals are able to taste this molecule while 1200 do not. a) What are the allele frequencies? b) What are the genotype frequencies? Solution: a) What are the allele frequencies? Assume that : G : able to taste (p) g : unable to taste (q) 1200 unable to taste => q^2 = 1200/(2800+1200) = 0.3 => q = sqrt(0.3) => p = 1 – sqrt(0.3) . b) What are the genotype frequencies? GG = p^2 = (1 – sqrt(0.3))^2 Gg = 2pq = 2sqrt(0.3)(1-sqrt(0.3)) gg = q^2 = 0.3 B. In humans, the Rh factor genetic information is inherited from our parents, but it is inherited independently of the ABO blood type alleles. In humans, Rh+ individuals have the Rh antigen on their red blood cells, while Rh− individuals do not. There are two different alleles for the Rh factor known as Rh+ and rh. Assume that a dominant gene Rh produces the Rh+ phenotype, and that the recessive rh allele produces the Rh− phenotype. In a population that is in Hardy-Weinberg equilibrium, if 190 out of 200 individuals are Rh+, calculate the frequencies of both alleles. Solution: Assume that : Rh+ : p ; Rh- : q ( allelic frequency) 190 out of 200 are Rh+ = > 10 out of 200 are Rh=> q^2 = 10/200 = 0.05 => q = sqrt(0.05) => p = 1- sqrt(0.05) 23 5. Premature baldness is a trait influenced by sex: it is dominant in men and recessive in women. In a study of 10000 men, 7225 were not bald. How many individuals are expected to have normal hair in a population of 10000 women? Solution: Assume that : H : Bald (p) h : Normal (q) Trait is influenced by sex => Hh in men : Bald ; Hh in women : Normal 10000 men , 7225 normal => hh = 7225/10000 = 0.7225 = q^2 => q = 0.85 => p = 1-0.85 = 0.15 In women ; Normal are : Hh + hh = 2x0.15x0.85 + 0.85^2 = 0.9775 => 9775 women are expected to have normal hair. 6. A. An allele W, for white wool, is dominant over allele w, for black wool in a sheep. In a sample of 900 sheep, 891 are white and 9 are black. Calculate the allelic frequencies within this population, assuming that the population is in H-W equilibrium. Solution: White allelic frequency = p ; black allelic frequency = q 9 are black => ww = 9/900 = 0.01 => q^2 = 0.01 => q= 0.1 => p = 0.9 B. In the United States, approximately one child in 10,000 is born with PKU (phenylketonuria), a syndrome that affects individuals homozygous for the recessive allele (aa). (a) Calculate the frequency of this allele in the population. (b) Calculate the frequency of the normal allele. (c) Calculate the percentage of carriers of the trait within the population. Solution: (a) Calculate the frequency of this allele in the population. Allelic frequency : Normal : p ; PKU : q. 1/10000 PKU => q^2 = 1/10000 => q = 0.01 (b) Calculate the frequency of the normal allele. p = 1-0.01 = 0.99 (c) Calculate the percentage of carriers of the trait within the population. A hereditary carrier (genetic carrier or just carrier), is a person or other organism that has inherited a recessive allele for a genetic trait or mutation but usually does not display that trait or show symptoms of the disease. Carriers of the trait : Aa = 2pq = 2x0.01x0.99 = 1.98 % 7. In cats, the gene for hair color is located on a non-homologous region of the X chromosome: the dominant allele for black hair is completely dominant over the recessive allele for tabby hair, individuals with the heterozygous genotype has calico hairs. . A cat population is in genetic equilibrium with 10% male tabby cats. Theoretically, if the population has 1000 cats, how many calico cats are expected ? Solution: 24 Asumme that XA : black, Xa: tabby In XY: % tabby cat =2x10%(% of male tabby cat in population)=20% % Xa=20%=> %XA=80% In XX: %XAXa=2x20%x80%=32% In population: % calio cat=32%:2=16% 160 calio cat 8. In sheep, allele D codes for horns, allele d for no horns; genotype Dd for horns in males and no horns in females. In a population in genetic equilibrium, sheep have 50% horns. All hornless sheep in this population are randomly mated to obtain F1. Theoretically, in F1, what is the ratio of hornless sheep that can be expected ? Solution: % sheep having horn=%DD+%Dd/2=50% %𝐷2 + %𝐷xx(1-%D)=1/2 => %D=50%=> %d=50% All hornless sheep in this population are randomly mated to obtain F1: 𝑃:(♂)100%dd x (♀)(50%Dd+25%dd) Gamates: (♂) 100%d x (♀) (1/3 D+ 2/3 d) 1 2 F1: 1/3 Dd +2/3 dd=> hornless sheep: 3 : 2 + 3=5/6 25