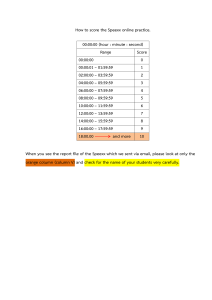
Programming for Biologist Part #1: SAMParser.py Next-generation sequencing technologies are making a large impact in Biology today. The ability to sequence large genomes and RNA molecules from cells in different conditions has allowed biologists to propose many different experiments. At the root of all the analysis the sequences have to be first aligned to the genome so you know where the sequence comes from. For this assignment you will parse the results of an alignment ( called a SAM/BAM file ) and compare it to a file that contains gene annotations (GTF) . The documentation for the SAM/BAM file can be found here. (https://samtools.github.io/htsspecs/SAMv1.pdf). The name of the BAM file we will be using is sample_sort.bam. The two columns of interest for our purpose are column 3, the name of the chromosome, and column 5, the start position of the alignment. Fortunately for us there is a library called PySAM (https://pysam.readthedocs.io/en/latest/) so we can parse the file easily. In order for us to use all the functionality of PySAM library, we need to also use an index file sample_sort.bam.bai. An index file is just like the index in a textbook, it tells the program where in the file the different chromosome’s alignments start. Now the GTF file (Arabidopsis.gtf) is a tab-separated file already sorted. It contains the chromosome in the first column and the start and end positions of an exon in the 4th and 5th column, and the gene name in the last column. Remember that for certain genes there are more than one transcripts. Which means exons are repeated. To resolve this we will use a simple rule, let’s just take the first transcript. So use only the exons that belong to the first transcript, their ID ends with a .1 To determine the number of reads that match a gene, count the number of start positions from the SAM file that fall between the start and stop coordinates of an exon that belongs to a gene. You can use the count function or the fetch function in Pysam (https://pysam.readthedocs.io/en/latest/api.html#pysam.AlignmentFile.count) to help you with this. Note - some reads can match multiple exons. To be technically correct, make sure you don’t count the same read twice. But for this assignment we will not deduct points for not keep tracking of reads that match more than one exon. The output should be a matrix with two columns, one for the gene name and the other for the number of reads that matched. Make sure to include all genes, even if they do not have a match. Part #2: Write your own KNN from scratch You are given a matrix where 20 genes are used as biomarkers to identify what type of cancer the patient has. This labeled data is training.txt. You are also given a smaller set of data (test.txt) for the same 20 genes, but you will have to predict the label ( the type of cancer). For each individual in the test set, use euclidean distance to find the closest k members of the individual in the Training set. Predict the label based on which label is in the majority among the labels of its neighbors. The output is the test set with the predicted labels. KNN.py will accept four arguments: 1. - r The training data 2. - t The test data 3. - k The k to be used 4. - o The name of the output where the predictions should be saved.