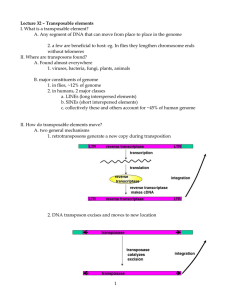
Genome organization in prokaryotes and eukaryotes Prof. Marcos De Donato, PhD DBI, Tecnológico de Monterrey Campus Querétaro Genome organization in prokaryotes and eukaryotes Prokaryotic Genomes 1. Genomes sequenced 2. Genome size and structure 3. Mobile elements Eukaryotic Genomes 1. Genome structure 2. Repetitive elements 3. Chromatin structure Prokaryotic Genomes Bacterial Cell Size Tree of Life Animals Plants The first phylogenetic tree based on rRNA data, emphasizing the separation of bacteria, archaea, and eukaryotes, as proposed by Woese et al. in 1990. Woese et al. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. PNAS, 1990; 87(12), 4576-4579. Tree of Life Animals Plants Pace NR. A molecular view of microbial diversity and the biosphere. Science, 1997; 276(5313):734-40. Modern Version of the Tree of Life A current view of the tree of life, encompassing the total diversity represented by sequenced genomes. The tree includes 92 named bacterial phyla, 26 archaeal phyla and all five of the Eukaryotic supergroups. Lineages lacking an isolated representative are shown with red dots. http://geschwindlab.neurology.ucla.edu/protocols/next-generation-sequencing Hug, Laura A., et al. A new view of the tree of life. Nature microbiology 1.5 (2016): 16048. All Known Animals All Known Plants Bacterial DNA http://myhome.sunyocc.edu/~weiskirl/parts_of_all_cells.htm http://fbio.uh.cu/sites/genmol/confs/conf1/p1.htm Bacterial Cell Viral Genome Sizes are Non-Randomly Distributed in the Tree of Life Campillo-Balderas et al. Frontiers in Ecology and Evolution. 2015 Dec 23;3:143. Core genome: Contains the genes that are essential to the basic functions of the species. These genes are called the housekeeping genes. Pangenome: Contains all the genes that can be found in any strain of the species (including those in the core genome). Strain specific genes are part of the pangenome, which can provide different functions that help the bacteria to adapt to different environments. Core vs Pangenomes Core vs Pangenomes There is a very large number of genes in the pan genome, compared to the number in the core genome of E. coli Core vs Pangenomes https://tel.archives-ouvertes.fr/tel-01599361/document Bacterial DNA Structure Prokaryotic Cell Plasmids Bacterial Chromosome https://www.shmoop.com/biology-cells/prokaryotic-cells.html Bacterial DNA Structure http://biology.kenyon.edu/courses/biol114/Chap01/chrom_struct.html Bacterial DNA Structure http://biology.kenyon.edu/courses/biol114/Chap01/chrom_struct.html Bacterial DNA Structure http://biology.kenyon.edu/courses/biol114/Chap01/chrom_struct.html Bacterial Mobile Elements Frost et al. Nature Reviews Microbiology. 2005; 3(9):722. Bacterial Mobile Elements Finan TM. Microbiol. Mol. Biol. Rev. 2017; 81(3):e00019-17. Eukaryotic Genomes Mitochondrial and Plastid Endosymbiosis Schematic tree of newer hypotheses for phylogenetic relationships among major groups of eukaryotes. Martin, W. & Mentel, M. (2010) The Origin of Mitochondria. Nature Education 3(9):58. Mitochondrial and Plastid Endosymbiosis Figure 1. Mitochondrion-late and mitochondrion-early models for the origin of eukaryotes. Fossil evidence has it that eukaryotes are 1.5 billion to 1.8 billion years old. All current models for the origin of eukaryotes have mitochondria in the eukaryote common ancestor. (A) In mitochondrion-late models, an archaeon (red) becomes a complex protoeukaryote, evolves phagocytosis, and acquires the proteobacterium (blue). (B) In mitochondrion-early models, phagocytosis came after the mitochondrion. Mitochondrionearly models typically start with metabolic interactions between an archaeon and the proteobacterial ancestor of mitochondria. Martin et al. Microbiol Mol Biol Rev. 2017;81(3). pii: e00008-17. Secondary Endosymbiosis Martin et al. Microbiol Mol Biol Rev. 2017;81(3). pii: e00008-17. Evolution of the Domains of Life López-García P, Eme L, Moreira D. J Theor Biol. 2017 Dec 7;434:20-33. C-Value Paradox https://www.mun.ca/biology/desmid/brian/BIOL2060/BIOL2060-18/CB18.html Genome Sizes in Viruses, Bacteria, Archaea and Eukaryotes Koonin, EV. 2011. The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press. Relation between Gene Fraction & Genome Size in Eukaryotes Drosohila A. thaliana https://slideplayer.com/slide/8850552/ Genome Size Variation in Amniotes Kapusta A, Suh A, Feschotte C. Dynamics of genome size evolution in birds and mammals. PNAS. 2017; 114(8):E1460-9. Different Components Making up the Human Genome About 1.5% of the genome consists of the ≈20,000 protein-coding sequences which are interspersed by the non coding introns, making up about 26%. Transposable elements are the largest fraction (40-50%) including for example long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs). Most transposable elements are genomic remnants, which are currently defunct. (Gregory TR. Nat Rev Genet. 9:699-708, 2005) Barbara McClintock and Her Jumping Genes 1940s, Barbara McClintock discovered the first transposable element in maize, earned a Nobel prize in 1983. Late 1960s, transposition was also found in Bacteria. http://newshub.agilent.com/2015/06/16/barbara-mcclintock-and-her-jumping-genes/ Aleurone Color in Maize Aleurone is a layer surrounding the maize seed and the color is determined by several genes. Only when the aleurone has no color (transparent), the color of the endosperm can be seen. Colorless Substance c/c C2 X Colorless Substance 2 pr1/pr1 Pr1 X Red Pigment Bz1 X bz1/bz1 Purple Pigment Ds/Ac Mobile Elements in Maize The movement of mobile elements can induce mutations including chromosome brakage. This is how McClintock found out about these elements in maize. The excision of the mobile element from the gene makes it functional again, allowing the production of pigment, but only in the cells that have inherited the functional copy (purple patches). Those cells where the gene is still disrupted by Ds will show the yellow background). http://www.bio.miami.edu/dana/250/250S12_17print.html Ds/Ac Mobile Elements in Maize Normal gene producing pigment Disrupted gene not producing pigment Cells where Ds have excise out of the gene, cells resume pigment production Ac can also disrupt the gene and when it excise out of the gene, cells resume pigment production http://www.bio.miami.edu/dana/250/250S12_17print.html Disruption of gene function by Mobile Elements Transposable elements can be inserted in different places within a gene, causing different effects. http://www.bx.psu.edu/~ross/workmg/TranspositionCh9.htm P Element in Drosophila P element in drosophila is a mobile element causing sterility when they become active. http://www.bio.miami.edu/dana/250/250S12_17print.html P Element in Drosophila In the fly gametes, the P element can become active, inserting into many places and causing disruption in several important genes, resulting in sterility. Only when the P element is inherited from the mother, the individuals also get suppressors of the P element movement, not allowing them to move. http://www.bio.miami.edu/dana/250/250S12_17print.html Types of Transposable Elements Sampath & Yang. Plant Breeding and Biotechnology. 2014; 2(4):322-33. Transposition in Transposable Elements (TEs) Levin & Moran. Nature Reviews Genetics, 2011; 12(9):615-627. Mobile DNA Elements https://openi.nlm.nih.gov/detailedresult.php?img=PMC3386198_ppat.1002790.g003&req=4 SINEs Short interspersed elements (SINEs) are defined as relatively short (< 700 bp) nonautonomous retroposons transcribed by the cellular RNA polymerase III (pol III) from an internal promoter, while their reverse transcription depends on the RT of partner LINEs. • Most are tRNA derived; Alu is 7SL-RNA • Nonautonomous • Dependent on other machinery- genome “parasite” • RNA Pol III • Needs LINE Endonuclease and Reverse Transcriptase for activity • Composed of 3 parts: 5’ head, Body, 3’ tail Average size ~200 base pairs Vassetzky & Kramerov. Nucleic acids research. 2012; 41(D1):D83-9: http://sines.eimb.ru/ Polymorphism of Alu Insertion The ancestral human population is shown at the top, and two separate subpopulations as shown here. A monomorphic Alu insertion (red) is shared by all members of the population. Several Alu insertion polymorphisms are also shown, including an intermediatefrequency Alu insertion polymorphism in the ancestral and subpopulations (green), a population-specific element (blue) and a de novo insert in subpopulation B (mauve). Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nature reviews genetics. 2002 May;3(5):370. The Impact of TEs on Mammalian Genomes Garcia-Perez JL, Widmann TJ, Adams IR. The impact of transposable elements on mammalian development. Development. 2016; 143(22):4101-14. Silencing Mechanisms Controlling TE Activity Horváth V, Merenciano M, González J. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends in Genetics. 2017; 33(11)832-41. Effect of Stress on TEs Horváth V, Merenciano M, González J. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends in Genetics. 2017; 33(11)832-41. TEs Can Alter Expression and Structure of Nearby Genes Horváth V, Merenciano M, González J. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends in Genetics. 2017; 33(11)832-41. Repetitive Elements Most of the human genome is made of repetitive elements, with the largest fraction originated from transposable and retrotransposable elements. LINEs and SINEs (Alus) are the most frequent retrotransposons. Exon Shuffling by Homologous Recombination Repetitive elements, like Alu elements can participate in a process called exon shuffling, where genes can exchange exons, changing the proteins they code, allowing for the creation of new functions. Exon Shuffling by Transposition Exon Shuffling Exon Shuffling can produce new genes with novel functions that can help the individual to adapt to new environmental conditions or niches. This is especially important when the genes have been previously duplicated, allowing for the original gene to conserve the original function and the duplicated gene to acquire the new function. Processed pseudogenes can re-acquire a function if they are inserted next to a promoter of another gene. DNA Organization in Eukaryotes DNA Organization in Eukaryotes CHROMOSOME STRUCTURE CHROMOSOME Telomeres STRUCTURE Arms Centromere Chromatids Telomeres "SMC" proteins- structural maintenance of chromosomes https://www.studyblue.com/notes/note/n/chapter-4-dna-chromosomes-and-genomes/deck/8763553 Telomeres http://www.sens.org/research/research-blog/2013-research-report-part-3-12-cancerous-cells Neumann & Reddel. Nature Rev Cancer. 2:879-884, 2002 Centromere Centromere structure and organization. (A) Centromeric chromatin underlies the kinetochore, which contains inner and outer plates that form microtubule-attachment sites. Pericentromeric heterochromatin flanks centromeric chromatin, and contains a high density of cohesin, which mediates sister-chromatid cohesion. (B) Schematic depiction of centromeric DNA in humans and mice, Drosophila, Schizosaccharomyces pombe, Candida albicans and Saccharomyces cerevisiae. Allshire & Karpen. Nature Rev Genet, 9:923-937, 2008