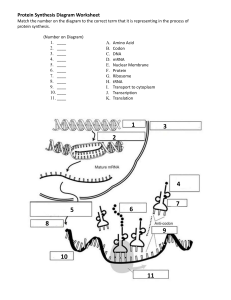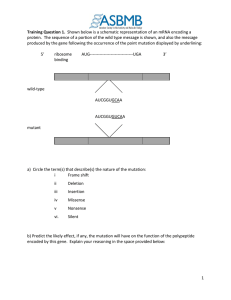
CH15: TRANSLATION mRNA stability • The steady-state level: the amount of mRNA in the cell • combination of transcription rate and rate of mRNA degradation • Determines the amount of mRNA available for translation • Regulated in response to cellular needs . RNAi gene silencing • RNA interference uses Dicer to cleave double-stranded RNA molecules into small interfering RNAs (siRNAs) and micro RNAs (miRNAs) • siRNAs and miRNAs repress mRNA translation and trigger degradation Why translate? Amino acids are joined together by peptide bonds. The Genetic Code: RNA to Amino Acids Second base of RNA codon UUU U UUC UUA First base of RNA codon UUG UCU Phenylalanine (Phe) UCC Leucine (Leu) CUU C CUC CUA A G C Leucine (Leu) A UAU Serine (Ser) UAC UAG Stop UGG Tryptophan (Trp) G CCU CAU CCC CCA Proline (Pro) CAC CAA ACU AAU ACC AUA ACA AAC Threonine (Thr) AAA AUG Met or start ACG AAG GUU GCU GAU GUA GUG C UCG AUU Valine (Val) U UGA Stop CAG GUC UGC Cysteine (Cys) Stop CCG Isoleucine (Ile) UGU UAA UCA CUG AUC Tyrosine (Tyr) G GCC GCA GCG Alanine (Ala) GAC GAA GAG Histidine (His) CGA Glutamine (Gln) CGG AGU Asparagine (Asn) AGC Lysine (Lys) U CGU CGC AGA AGG GGU Aspartic acid (Asp) GGC GGA Glutamic acid (Glu) GGG A Arginine (Arg) C A G Serine (Ser) Arginine (Arg) U C A G U Glycine (Gly) C A G Third base of RNA codon U . Figure 14.1 tRNA • tRNAs are small in size and very stable • • Composed of 75–90 nucleotides Transcribed from DNA and contain post-transcriptionally modified bases • • Modified bases enhance hydrogen bonding efficiency during translation The two-dimensional structure of tRNAs is a cloverleaf Figure 14.3 Wobble may exist in the pairing of a codon and anticodon. The mRNA and tRNA pair in an antiparallel fashion. Proteins in 3 “Easy” Steps!! • Initiation • Elongation • Termination Initiation Met Large ribosomal subunit Initiator tRNA P site mRNA 2 1 Start codon Small ribosomal subunit A site Prokaryotes: Shine-Dalgarno Sequence The poly(A) tail of eukaryotic mRNA plays a role in the initiation of translation. Eukaryotes Elongation • The charged tRNAs enter the A site, and peptidyl transferase catalyzes peptide bond formation • The uncharged tRNA moves to the E (exit) site • The tRNA bound to the peptide chain moves to the P site • The sequence of elongation and translocation is repeated over and over Termination • Termination: signaled by a stop codon (UAG, UAA, UGA) in the A site • GTP-dependent release factors cleave the polypeptide chain from the tRNA and release it Figure 14.17 Figure 14.18 Figure 14.19 Figure 14.20 Post-translational modifications